
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,913,334 – 19,913,436 |
| Length | 102 |
| Max. P | 0.546973 |

| Location | 19,913,334 – 19,913,436 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.62 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -15.50 |
| Energy contribution | -14.62 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19913334 102 + 20766785 UGACCCUAU----CCC---AUCUCCUCACCAGCAGGAACACAGGGCUGAUGUACGUCCAGCAGAUCCGCCAGAAUAAGCCUGGCUUGCUGCCCAGCAUCUGUUCCACAU .........----...---...............((((((..((((........)))).((((....(((((.......)))))...))))........)))))).... ( -26.80) >DroVir_CAF1 3121 95 + 1 UAAA----------CU---GU-CACCUACCAGCAAGAAUACUGGACUUAUAUAGGUCCAACAGAUGCGCCAGAAGAUGCCCGGCUUAGAGCCCAACAUUUGCUCCACAU ....----------((---((-.(((((((((........)))).......)))))...))))(((.(..((.(((((...(((.....)))...))))).)).).))) ( -17.81) >DroYak_CAF1 2551 106 + 1 UAACCCAUCUUCUUUU---UUCUUCUCACCAGCAGGAACACGGGACUAAUGUACGUCCAGCAGAUCCGCCAGAACAGGCCUGGCUUGCUGCCCAGCAUCUGUUCCACAU ................---...............((((((..((((........)))).((((....(((((.......)))))...))))........)))))).... ( -27.20) >DroMoj_CAF1 8844 96 + 1 CUCU----------CU---GUCCACUUACCAGCAGAAACACGGGGCUUAUGUACGUCCAACAGAUGCGCCAGAAAAUCCCCGGCUUGGCGCCCAGCAUCUGCUCCACAU ...(----------((---((..........))))).....(((((..((((..(((.....)))(((((((............)))))))...))))..))))).... ( -25.70) >DroAna_CAF1 2483 104 + 1 UAAA-AUAU----UCCUUAGUUUACUUACCAGCAGGAACACGGGACUGAUGUAUGUCCAGCAUAUGCGCCAGAAUAUUCCCGGCUUAGCUCCCAGCAUCUGUUCUACAU ....-....----.....................((((((((((((((.(((((((.....))))))).))).....)))))(((........)))...)))))).... ( -22.10) >DroPer_CAF1 2557 86 + 1 -----------------------UCUUACCAGCAGGAACACCGGACUGAUGUAGGUCCAGCAGAUGCGCCAGAACAGUCCCGGCUUGGAGCCCAGCAUCUGCUCCACAU -----------------------...........(((.....(((((......))))).(((((((((((.((....))..))).(((...)))))))))))))).... ( -29.90) >consensus UAAA__________CU___GUCUACUUACCAGCAGGAACACGGGACUGAUGUACGUCCAGCAGAUGCGCCAGAACAUGCCCGGCUUGGAGCCCAGCAUCUGCUCCACAU ..............................((((((......((((........)))).........(((.(.......).))).............))))))...... (-15.50 = -14.62 + -0.89)

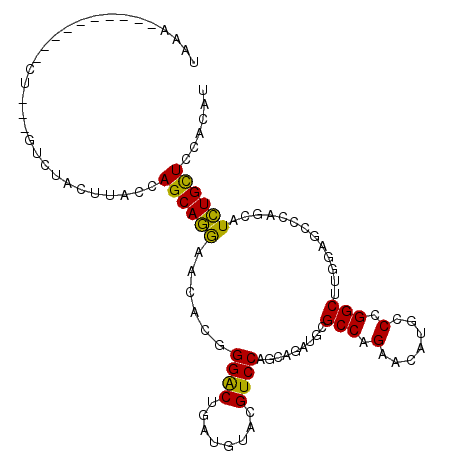
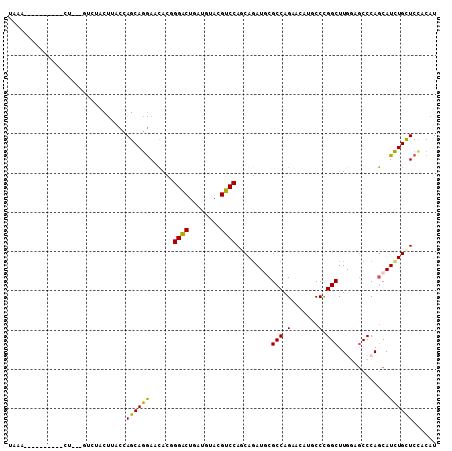
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:55 2006