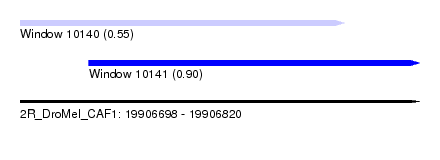
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,906,698 – 19,906,820 |
| Length | 122 |
| Max. P | 0.904994 |

| Location | 19,906,698 – 19,906,797 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.60 |
| Mean single sequence MFE | -36.01 |
| Consensus MFE | -31.88 |
| Energy contribution | -31.79 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
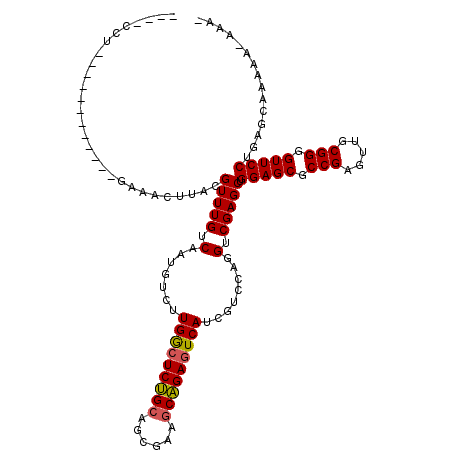
>2R_DroMel_CAF1 19906698 99 + 20766785 U-------------------AUGCUUACGUUUGUCAAUGUCUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGAGAAAGAU-UAA- .-------------------...((((.(((((.(.(((...(((((((((......))))))))).)))....).))))).(((((.(((.....))).)))))))))......-...- ( -34.50) >DroVir_CAF1 184205 102 + 1 ----GCU------------UGCACUUACGUUUGUCAAUGUCUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGGCCAAGAA-AAA- ----(((------------((.(((((((((....)))))..(((((((((......)))))))))......))))))))).(((((.(((.....))).)))))..........-...- ( -36.70) >DroPse_CAF1 146646 106 + 1 UAUGCCU------------UCAACUUACGUUUGUCAAUGUCUUGGCUCCGCAGAGAGGCGGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUAAUGGAAAU-CAA- ....(((------------.((((((.(((............(((((((((......)))))))))...(((((......))))))))..)))))).))).((((....))))..-...- ( -38.00) >DroGri_CAF1 183461 114 + 1 ----ACUCAUCACUCAAGUGCAACUUACGUUUGUCAAUGUCUUGGCUCUGCAGCGAGGCAGAAUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGUGCAAGAA-AGU- ----.......(((....((((....(((((....)))))...(((((((((((..(((.((....)).(((((......)))))..)))..)))))))))))....))))....-)))- ( -33.90) >DroWil_CAF1 312101 108 + 1 UAUGCUC------------CAUGCUUACGUUUGUCAAUAUCUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGGUUAAAAAAGACA ......(------------((.......(((((.(.......(((((((((......)))))))))........).))))).(((((.(((.....))).))))))))............ ( -34.76) >DroMoj_CAF1 233944 102 + 1 ----CUG------------GACACUUACGUUUGUCAAUGUCUUGACUCUGCAGCGAAACAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUAAGCAAAAG-AAA- ----(((------------(((....(((((....)))))..((((((((........))))))))..)))))).....((((((((.(((.....))).)))))..))).....-...- ( -38.20) >consensus ____CCU____________GAAACUUACGUUUGUCAAUGUCUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGAGCAAAAA_AAA_ ............................(((((.(.......(((((((((......)))))))))........).))))).(((((.(((.....))).)))))............... (-31.88 = -31.79 + -0.08)

| Location | 19,906,719 – 19,906,820 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.11 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -27.00 |
| Energy contribution | -27.20 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
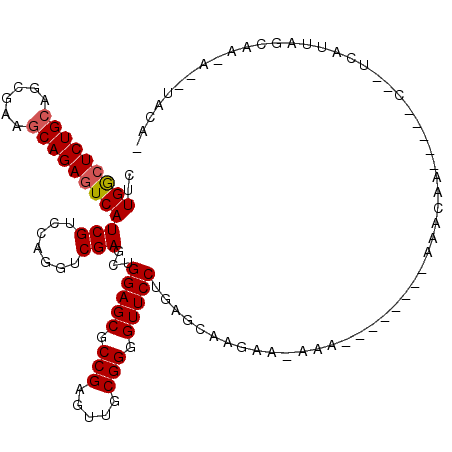
>2R_DroMel_CAF1 19906719 101 + 20766785 CUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGAGAAAGAU-UAA--------AUACAG-----C--UUAUUAGCAU-A--UUCAU ..(((((((((......))))))))).......((((...(((((((.(((.....))).)))))..))...)))-)..--------.....(-----(--(....)))..-.--..... ( -32.90) >DroVir_CAF1 184229 102 + 1 CUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGGCCAAGAA-AAA--------AGAUGG-----CUA-CGUUAGUAAAA--UACC- ..(((((((((......)))))))))(((((....((..((((((((.(((.....))).))))).)))...)).-...--------.)))))-----.((-(....)))...--....- ( -35.80) >DroGri_CAF1 183497 102 + 1 CUUGGCUCUGCAGCGAGGCAGAAUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGUGCAAGAA-AGU--------AAAAUA-----CAAUCAUUAGCAA-A--UACU- ...(((((((((((..(((.((....)).(((((......)))))..)))..)))))))))))....(((((((.-.((--------(...))-----)..)).)).))).-.--....- ( -29.90) >DroWil_CAF1 312129 108 + 1 CUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGGUUAAAAAAGACAGACAGAGAGACAA-----C--AAAUUAGUAU-----UACU ..(((((((((......)))))))))..(((...(((.(((((((((.(((.....))).))))).))))......))).)))..........-----.--..........-----.... ( -35.70) >DroMoj_CAF1 233968 103 + 1 CUUGACUCUGCAGCGAAACAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUAAGCAAAAG-AAA-C------ACAACA-----UAA-UGUCAGUAAAA--UACA- ..((((((((........))))))))(((.......)))((((((((.(((.....))).)))))..))).....-...-.------......-----...-...........--....- ( -28.20) >DroAna_CAF1 135479 105 + 1 CUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCU---AAAGGU-GAG--------AAACGGAAAUCC--UCGUUAGCAC-UUGUUUUU ..(((((((((......)))))))))(((.......)))...(((((.(((.....))).))))).---..((((-(..--------.(((((......--)))))..)))-))...... ( -36.40) >consensus CUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGAGCAAGAA_AAA________AAACAA_____C__UCAUUAGCAA_A__UACA_ ..(((((((((......)))))))))(((.......)))...(((((.(((.....))).)))))....................................................... (-27.00 = -27.20 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:50 2006