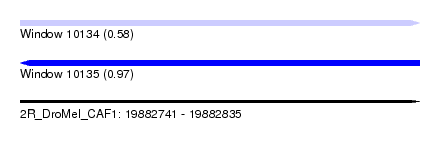
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,882,741 – 19,882,835 |
| Length | 94 |
| Max. P | 0.968373 |

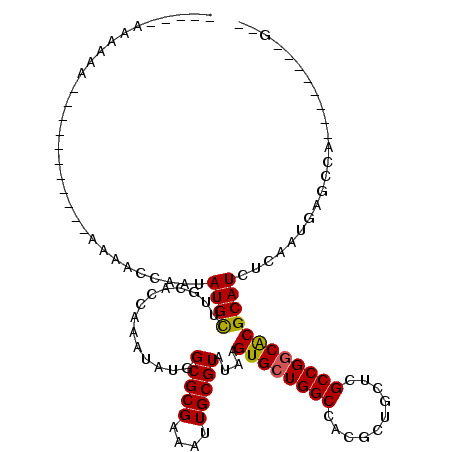
| Location | 19,882,741 – 19,882,835 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.22 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -15.80 |
| Energy contribution | -16.05 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19882741 94 + 20766785 --C--------CAGCUCAUUGAGAUGCGUGCCGGCAAACAGCCUGGCCAGCACUUAUACGCAAUUUCGCGCCAUAUUUGGUUCAAACAUAUUGGAUUU-----------UUUUGU----- --(--------((((....((((.(((..((((((.....))).)))..)))))))...))......(.((((....)))).)........)))....-----------......----- ( -24.20) >DroVir_CAF1 155593 94 + 1 AGUUGCGCUCUUGGCUCAUUGAAAUGCGUGCCGGCGUGCAACGCGGCCAGCACUUAUACGCAAUUUCGCGCCAUAUUUGGUGCAAGCAUAUUGC-------------------------- .(((((((.((.(((.(((....)))...))))).)))))))((.....((........))......((((((....))))))..)).......-------------------------- ( -30.80) >DroGri_CAF1 156394 102 + 1 AGC--------UGGCUCAUUGAAAUGCGCGCCGGCGUGCAACGCGGCCAGCACUUAUACGCAAUUUCGCGCCAUAUUUGGUGCAAGCAUAUUGCUUUUUUCU-----UAUUUUUC----- .((--------(((((....(...((((((....))))))...))))))))........(((((...((((((....))))))......)))))........-----........----- ( -34.50) >DroWil_CAF1 257527 93 + 1 -----------AAGCUCAUUGAGAUGCGUGGCGGCUAACAGCCUGGCCAGCACUUAUACGCAAUUUCGCGCCAUAUUUGGAGCAAACAUAUUGGUUUU-----------UUUUUU----- -----------..((((..((((.(((.(((((((.....)))..)))))))))))..(((......))).........))))...............-----------......----- ( -25.40) >DroMoj_CAF1 205527 115 + 1 AGCGCUGCUCAUGGCUCAUUGAAAUGCGUGCCGGCGAGCAACGCGGCCAGCACUUAUACGCAAUUUCGCGCCAUAUUUGGUCCAAGCAUAUUGUUUUUCCCUCUUUAUAUUUUGU----- .(((.(((((.((((.(((....)))...))))..))))).)))((((((....(((.(((......)))..))).)))))).((((.....))))...................----- ( -28.20) >DroAna_CAF1 110812 99 + 1 --C--------CAUCUCAUUGAGAUGCGUGCCGGCAAACAGCCUGGCCAGCACUUAUACGCAAUUUCGCGCCAUAUUUGGUUCAAACAUAUUGGCUUU-----------UUUUUCGUACU --.--------((((((...)))))).((((.(((.....))).(((((((........))......(.((((....)))).)........)))))..-----------......)))). ( -25.00) >consensus __C________UAGCUCAUUGAAAUGCGUGCCGGCGAACAACCCGGCCAGCACUUAUACGCAAUUUCGCGCCAUAUUUGGUGCAAACAUAUUGGUUUU___________UUUUUU_____ ....................(((((((((((.(((..........))).))......)))).)))))(.((((....)))).)..................................... (-15.80 = -16.05 + 0.25)

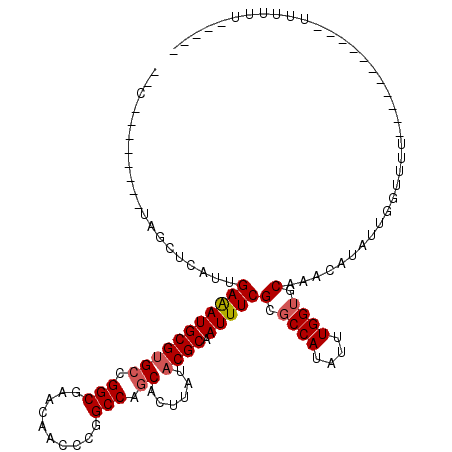
| Location | 19,882,741 – 19,882,835 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.22 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -20.47 |
| Energy contribution | -20.25 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19882741 94 - 20766785 -----ACAAAA-----------AAAUCCAAUAUGUUUGAACCAAAUAUGGCGCGAAAUUGCGUAUAAGUGCUGGCCAGGCUGUUUGCCGGCACGCAUCUCAAUGAGCUG--------G-- -----......-----------....(((.((((((((...))))))))(((((....)))))....((((((((..........))))))))((.((.....))))))--------)-- ( -24.90) >DroVir_CAF1 155593 94 - 1 --------------------------GCAAUAUGCUUGCACCAAAUAUGGCGCGAAAUUGCGUAUAAGUGCUGGCCGCGUUGCACGCCGGCACGCAUUUCAAUGAGCCAAGAGCGCAACU --------------------------((.((((((((((.(((....))).))))....))))))..((((((((.((...))..)))))))))).......((.((.....)).))... ( -31.00) >DroGri_CAF1 156394 102 - 1 -----GAAAAAUA-----AGAAAAAAGCAAUAUGCUUGCACCAAAUAUGGCGCGAAAUUGCGUAUAAGUGCUGGCCGCGUUGCACGCCGGCGCGCAUUUCAAUGAGCCA--------GCU -----........-----...........((((((((((.(((....))).))))....))))))....(((((((((((((.....)))))))(((....))).))))--------)). ( -31.10) >DroWil_CAF1 257527 93 - 1 -----AAAAAA-----------AAAACCAAUAUGUUUGCUCCAAAUAUGGCGCGAAAUUGCGUAUAAGUGCUGGCCAGGCUGUUAGCCGCCACGCAUCUCAAUGAGCUU----------- -----......-----------...............((((........(((((....)))))....((((((((..(((.....))))))).))))......))))..----------- ( -24.20) >DroMoj_CAF1 205527 115 - 1 -----ACAAAAUAUAAAGAGGGAAAAACAAUAUGCUUGGACCAAAUAUGGCGCGAAAUUGCGUAUAAGUGCUGGCCGCGUUGCUCGCCGGCACGCAUUUCAAUGAGCCAUGAGCAGCGCU -----................................((.(((.((...(((((....)))))....))..)))))((((((((((..(((...(((....))).))).)))))))))). ( -33.00) >DroAna_CAF1 110812 99 - 1 AGUACGAAAAA-----------AAAGCCAAUAUGUUUGAACCAAAUAUGGCGCGAAAUUGCGUAUAAGUGCUGGCCAGGCUGUUUGCCGGCACGCAUCUCAAUGAGAUG--------G-- .(((((.....-----------...((((...((((((...)))))))))).........)))))..((((((((..........)))))))).((((((...))))))--------.-- ( -31.63) >consensus _____AAAAAA___________AAAACCAAUAUGCUUGCACCAAAUAUGGCGCGAAAUUGCGUAUAAGUGCUGGCCACGCUGCUCGCCGGCACGCAUCUCAAUGAGCCA________G__ ...............................((((..............(((((....)))))....((((((((..........))))))))))))....................... (-20.47 = -20.25 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:45 2006