| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,870,637 – 19,870,739 |
| Length | 102 |
| Max. P | 0.933885 |

| Location | 19,870,637 – 19,870,739 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -21.76 |
| Energy contribution | -23.32 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19870637 102 + 20766785 -------CUUCGCCGUUGUGCAAC-GACUUUAAAAAGCUGUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACAGGCGACUCCUGGUCGCCACCAGCAGCCGUAC -------..(((((((((((((((-(.........(((((.((((....)))).))..)))......))))))))))).)))))((.(((((....))))).))...... ( -34.06) >DroGri_CAF1 139199 82 + 1 -------UGUUGUUGUUGUGCAAUUGCCUUUAAAAAGCUGUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACGCG--ACGC----GCAG--------------- -------.(((((.((((((((((...........(((((.((((....)))).))..))).......)))))))))))))--))..----....--------------- ( -22.17) >DroSim_CAF1 81133 102 + 1 -------CUCCGGCGUUGUGCAAC-GACUUUAAAAAGCUGUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACAGGCGACUCCUGGUCGCCACCAGCAGCCGUAC -------...((((((((((((((-(.........(((((.((((....)))).))..)))......))))))))))).((((((.....)))))).......))))... ( -38.76) >DroEre_CAF1 106298 109 + 1 CCAUUGGUUCCAGCGUUGUGCAAC-GACUUUAAAAAGCUGUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACAGGCGACUCCUGGUCGCCACCAGCAGCCGUAC .....((((...((((((((((((-(.........(((((.((((....)))).))..)))......))))))))))).((((((.....))))))....)))))).... ( -36.56) >DroYak_CAF1 104411 102 + 1 -------UUGCAGCGUUGUGCAAC-GACUUUAAAAAGCUGUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACAGGCGACUCCUGGUCGCCACCAGCAGCCGUAC -------.(((.((((((((((((-(.........(((((.((((....)))).))..)))......))))))))))).((((((.....)))))).......)).))). ( -35.46) >DroMoj_CAF1 189615 89 + 1 --AU---UGUUGCAGUUGUGCAAUUGCCUUUAAAAAGCUAUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACGCA--UCGC----GGCGCGGCG---------- --..---.(((((.((((((((((...........(((...((((....)))).....))).......))))))))))((.--....----.))))))).---------- ( -24.67) >consensus _______UUUCGGCGUUGUGCAAC_GACUUUAAAAAGCUGUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACAGGCGACUCCUGGUCGCCACCAGCAGCCGUAC ..............((((((((((...........(((((.((((....)))).))..))).......)))))))))).((((((.....)))))).............. (-21.76 = -23.32 + 1.56)

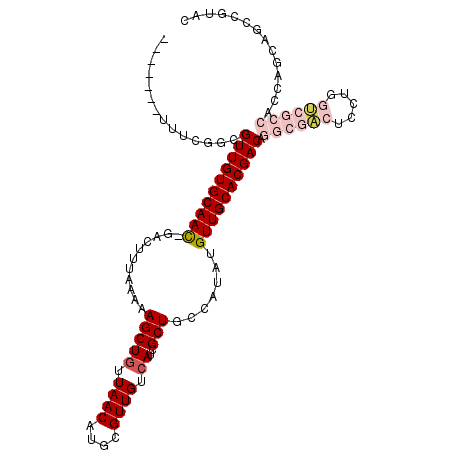
| Location | 19,870,637 – 19,870,739 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -21.14 |
| Energy contribution | -24.02 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19870637 102 - 20766785 GUACGGCUGCUGGUGGCGACCAGGAGUCGCCUGUCGUGCAACAUAUGGCAGCGAUGACAACGCAUGUUAACAGCUUUUUAAAGUC-GUUGCACAACGGCGAAG------- ....((((.(((((....))))).))))(((.((.(((((((...((((((((.......))).)))))...((((....)))).-))))))).)))))....------- ( -38.70) >DroGri_CAF1 139199 82 - 1 ---------------CUGC----GCGU--CGCGUCGUGCAACAUAUGGCAGCGAUGACAACGCAUGUUAACAGCUUUUUAAAGGCAAUUGCACAACAACAACA------- ---------------.(((----((((--.(((((((((........))).))))).).)))).........((((.....))))....)))...........------- ( -21.20) >DroSim_CAF1 81133 102 - 1 GUACGGCUGCUGGUGGCGACCAGGAGUCGCCUGUCGUGCAACAUAUGGCAGCGAUGACAACGCAUGUUAACAGCUUUUUAAAGUC-GUUGCACAACGCCGGAG------- ...((((.......((((((.....)))))).((.(((((((...((((((((.......))).)))))...((((....)))).-))))))).))))))...------- ( -37.30) >DroEre_CAF1 106298 109 - 1 GUACGGCUGCUGGUGGCGACCAGGAGUCGCCUGUCGUGCAACAUAUGGCAGCGAUGACAACGCAUGUUAACAGCUUUUUAAAGUC-GUUGCACAACGCUGGAACCAAUGG ....((...(..((((((((.....)))))).((.(((((((...((((((((.......))).)))))...((((....)))).-))))))).))))..)..))..... ( -36.20) >DroYak_CAF1 104411 102 - 1 GUACGGCUGCUGGUGGCGACCAGGAGUCGCCUGUCGUGCAACAUAUGGCAGCGAUGACAACGCAUGUUAACAGCUUUUUAAAGUC-GUUGCACAACGCUGCAA------- (((((((....((((((........)))))).)))))))........((((((.((.(((((.((.((((.......)))).)))-))))..)).))))))..------- ( -37.00) >DroMoj_CAF1 189615 89 - 1 ----------CGCCGCGCC----GCGA--UGCGUCGUGCAACAUAUGGCAGCGAUGACAACGCAUGUUAAUAGCUUUUUAAAGGCAAUUGCACAACUGCAACA---AU-- ----------......(((----(((.--((((((((((........))).))))).)).)))...((((.......)))).)))..(((((....)))))..---..-- ( -23.90) >consensus GUACGGCUGCUGGUGGCGACCAGGAGUCGCCUGUCGUGCAACAUAUGGCAGCGAUGACAACGCAUGUUAACAGCUUUUUAAAGUC_GUUGCACAACGCCGAAA_______ ...(((((.(((((....))))).)))))...((.(((((((...((((((((.......))).)))))...((((....))))..))))))).)).............. (-21.14 = -24.02 + 2.88)

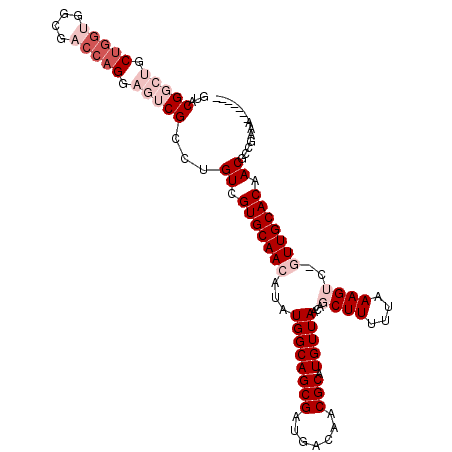
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:42 2006