
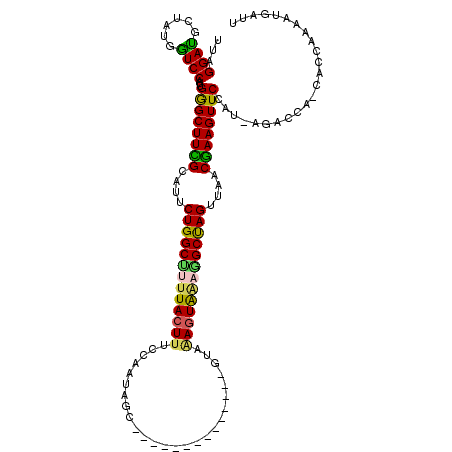
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,856,418 – 19,856,522 |
| Length | 104 |
| Max. P | 0.999738 |

| Location | 19,856,418 – 19,856,522 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.72 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -27.03 |
| Energy contribution | -26.39 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.44 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.74 |
| SVM decision value | 3.98 |
| SVM RNA-class probability | 0.999738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19856418 104 + 20766785 UUAGGAUGCUAUGAUCCACUGGGCUUCGCCUUCUGGCUUUUACUUGCCACUAGC--------------GUAAAGUACAGGCUAGUUAACGAAGUUCCAU-AGACCA-CACCAAAAUGAAU ...((...(((((.......((((((((....(((((((.(((((..(......--------------)..))))).)))))))....)))))))))))-)).)).-............. ( -27.61) >DroSec_CAF1 98403 104 + 1 UUAGGACGCUAUGGUCCACUGGGCUUCGCAAUCUGGCCUUUACUUUCCAAUAGC--------------GUAAAGUAGAGGCUAGUUAACGAAGUUCCAU-AGACCA-CACCAAAAUAAUU ...........(((((...(((((((((....((((((((((((((.(......--------------).))))))))))))))....))))).)))).-.)))))-............. ( -37.40) >DroSim_CAF1 75276 104 + 1 UUAGGAUGCUAUGGUCCACGGAGCUUCGCACUCUGGCUUUUACUUUCCAAUAGC--------------GCUUAUUGGAGGCUAGUUAACGAAGUUCCAU-AGACCA-CACCAAAAUGAUU ...((.((...(((((...(((((((((....((((((.......((((((((.--------------..))))))))))))))....)))))))))..-.)))))-))))......... ( -35.61) >DroEre_CAF1 100290 119 + 1 CUGGGAUGCUGUGGUCCACUGGGCUUUGUAUUCUGGCUCUUACUUUCCAAACGCAUAGAACGGCGAUUGUAGAGUAACUGCCAGCUUGCGAAGUUCCAUUAGAACA-CUCCGAAGUGAUU (((((((......))))..(((((((((((..(((((..(((((((.(((.(((........))).))).)))))))..)))))..))))))).)))).)))..((-((....))))... ( -41.40) >DroYak_CAF1 98256 115 + 1 CUAGGAUGCUAUGGUCCACCGGGCUUUGUAUUCUGGCGCUUACUUUACAAAAGUUAA-----AUGAUUGUAGAGUAACUGCUAGCUUACAAAGUUCAAAUGGAACCCCUCCAAAAUGACU ...(((.(....((((((..((((((((((..((((((.(((((((((((.......-----....))))))))))).))))))..))))))))))...))).)))).)))......... ( -39.50) >consensus UUAGGAUGCUAUGGUCCACUGGGCUUCGCAUUCUGGCUUUUACUUUCCAAUAGC______________GUAAAGUAAAGGCUAGUUAACGAAGUUCCAU_AGACCA_CACCAAAAUGAUU ...((((......))))...((((((((....(((((((((((((..........................)))))))))))))....))))))))........................ (-27.03 = -26.39 + -0.64)

| Location | 19,856,418 – 19,856,522 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.72 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -13.01 |
| Energy contribution | -13.93 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.38 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19856418 104 - 20766785 AUUCAUUUUGGUG-UGGUCU-AUGGAACUUCGUUAACUAGCCUGUACUUUAC--------------GCUAGUGGCAAGUAAAAGCCAGAAGGCGAAGCCCAGUGGAUCAUAGCAUCCUAA ........((.((-((((((-((((..(((((((.((((((.((.....)).--------------))))))(((........)))....))))))).))).))))))))).))...... ( -34.70) >DroSec_CAF1 98403 104 - 1 AAUUAUUUUGGUG-UGGUCU-AUGGAACUUCGUUAACUAGCCUCUACUUUAC--------------GCUAUUGGAAAGUAAAGGCCAGAUUGCGAAGCCCAGUGGACCAUAGCGUCCUAA .........(.((-((((((-((((..((((((...((.((((.((((((.(--------------(....)).)))))).)))).))...)))))).))).))))))))).)....... ( -37.30) >DroSim_CAF1 75276 104 - 1 AAUCAUUUUGGUG-UGGUCU-AUGGAACUUCGUUAACUAGCCUCCAAUAAGC--------------GCUAUUGGAAAGUAAAAGCCAGAGUGCGAAGCUCCGUGGACCAUAGCAUCCUAA ........((.((-((((((-(((((.((((((...((.((.(((((((...--------------..)))))))........)).))...)))))).))))))))))))).))...... ( -38.90) >DroEre_CAF1 100290 119 - 1 AAUCACUUCGGAG-UGUUCUAAUGGAACUUCGCAAGCUGGCAGUUACUCUACAAUCGCCGUUCUAUGCGUUUGGAAAGUAAGAGCCAGAAUACAAAGCCCAGUGGACCACAGCAUCCCAG .....(....).(-((.((((.(((..(((.(....(((((..(((((...(((.(((........))).)))...)))))..)))))....).))).))).)))).))).......... ( -31.00) >DroYak_CAF1 98256 115 - 1 AGUCAUUUUGGAGGGGUUCCAUUUGAACUUUGUAAGCUAGCAGUUACUCUACAAUCAU-----UUAACUUUUGUAAAGUAAGCGCCAGAAUACAAAGCCCGGUGGACCAUAGCAUCCUAG .........(((..(((.((((..(..(((((((..((.((..(((((.(((((....-----.......))))).)))))..)).))..))))))).)..)))))))......)))... ( -29.70) >consensus AAUCAUUUUGGUG_UGGUCU_AUGGAACUUCGUUAACUAGCCUCUACUCUAC______________GCUAUUGGAAAGUAAAAGCCAGAAUGCGAAGCCCAGUGGACCAUAGCAUCCUAA .........((...((((((..(((..((((((...((.((...((((............................))))...)).))...)))))).)))..))))))......))... (-13.01 = -13.93 + 0.92)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:40 2006