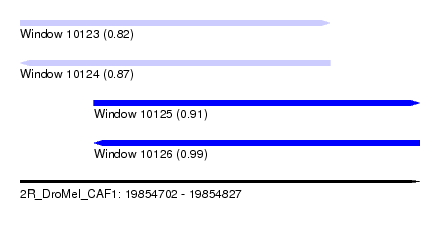
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,854,702 – 19,854,827 |
| Length | 125 |
| Max. P | 0.990222 |

| Location | 19,854,702 – 19,854,799 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -20.08 |
| Energy contribution | -21.08 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19854702 97 + 20766785 AUCGACACCUACUCAGAACCAGC------------AUCUGAAUCAGAAUCCGAAUGCGAUUUCGAAUCGGAAUCAGAACGAGAAUAAUCGGAAUCCGAUUCAGAUUCCG .......................------------(((((((((.((.(((((...((....))..))))).))....(((......)))......))))))))).... ( -24.20) >DroSec_CAF1 96702 97 + 1 AUCGACACCUACUCAGAAGCAGC------------AUCAGCAUCAGAAUCCGAAUCCGAUUUCGAAUCGGAAUCAGAACGAGAAUAAUCGGAAUCCGAUUCAGAUUCCG ...............((.((...------------....)).)).(((((.(((((.((((((((.(((.........)))......)))))))).))))).))))).. ( -23.80) >DroSim_CAF1 73584 97 + 1 AUCGACACCUACUCAGAAGCAGC------------AUCAGAAUCAGAAUCCGAAUCCGAUUUCGAAUCGGAAUCAGAACGAGAAUAAUCGGAAUCCGAUUCAGAUUCCG .......................------------....(((((.(((((.((.(((((((.....(((.........)))....))))))).)).))))).))))).. ( -24.60) >DroEre_CAF1 98489 109 + 1 AUCGACACCUACUCAGAACCAGACGCAGACGCAGACUCAGACUCAGAAUCAGAAUCCGAUUUUGAAUCGGAAUCAGAACGAGAAUAAUCGGAAUCCGAUUCAGAUUCCG ...........(((..........((....)).((.((.((.((((((((.......)))))))).)).)).)).....)))......(((((((.......))))))) ( -24.20) >DroYak_CAF1 96520 109 + 1 AUCGACACCUACUCAGAACCAGAACCAGAACCAGAAGCAGCAUCAGAAUCAGAAUCCGAUUUCGAAUCGGAAUCAGAACGAGAAUAAUCGGAAUACGAUUCAGAUUCCG .............................................(((((.(((((.(((((((...)))))))....(((......)))......))))).))))).. ( -18.20) >consensus AUCGACACCUACUCAGAACCAGC____________AUCAGAAUCAGAAUCCGAAUCCGAUUUCGAAUCGGAAUCAGAACGAGAAUAAUCGGAAUCCGAUUCAGAUUCCG .......................................(((((.(((((.((.(((((((.....(((.........)))....))))))).)).))))).))))).. (-20.08 = -21.08 + 1.00)

| Location | 19,854,702 – 19,854,799 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -23.13 |
| Energy contribution | -23.93 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19854702 97 - 20766785 CGGAAUCUGAAUCGGAUUCCGAUUAUUCUCGUUCUGAUUCCGAUUCGAAAUCGCAUUCGGAUUCUGAUUCAGAU------------GCUGGUUCUGAGUAGGUGUCGAU ((((((((((((((((.((.((..........)).)).))))))))(((......)))))))))))(((((((.------------......))))))).......... ( -30.80) >DroSec_CAF1 96702 97 - 1 CGGAAUCUGAAUCGGAUUCCGAUUAUUCUCGUUCUGAUUCCGAUUCGAAAUCGGAUUCGGAUUCUGAUGCUGAU------------GCUGCUUCUGAGUAGGUGUCGAU ((((((((((((((((...(((......))).))))).((((((.....)))))))))))))))))....((((------------((((((....)))).)))))).. ( -32.90) >DroSim_CAF1 73584 97 - 1 CGGAAUCUGAAUCGGAUUCCGAUUAUUCUCGUUCUGAUUCCGAUUCGAAAUCGGAUUCGGAUUCUGAUUCUGAU------------GCUGCUUCUGAGUAGGUGUCGAU .((((((.(((((.((.(((((((....(((.((.......))..)))))))))).)).))))).))))))(((------------((((((....)))).)))))... ( -33.50) >DroEre_CAF1 98489 109 - 1 CGGAAUCUGAAUCGGAUUCCGAUUAUUCUCGUUCUGAUUCCGAUUCAAAAUCGGAUUCUGAUUCUGAGUCUGAGUCUGCGUCUGCGUCUGGUUCUGAGUAGGUGUCGAU ..(((((.((((((((...(((......))).)))))))).)))))....((((((((.......))))))))(((.((..((((.((.......))))))..)).))) ( -37.00) >DroYak_CAF1 96520 109 - 1 CGGAAUCUGAAUCGUAUUCCGAUUAUUCUCGUUCUGAUUCCGAUUCGAAAUCGGAUUCUGAUUCUGAUGCUGCUUCUGGUUCUGGUUCUGGUUCUGAGUAGGUGUCGAU (((((((.(((.((...............))))).)))))))..((((.((((((.......)))))).((((((..((..((......))..))))))))...)))). ( -27.06) >consensus CGGAAUCUGAAUCGGAUUCCGAUUAUUCUCGUUCUGAUUCCGAUUCGAAAUCGGAUUCGGAUUCUGAUUCUGAU____________GCUGGUUCUGAGUAGGUGUCGAU (((((((.((((.(((..........))).)))).)))))))..((((.(((..(((((((..(.........................)..))))))).))).)))). (-23.13 = -23.93 + 0.80)

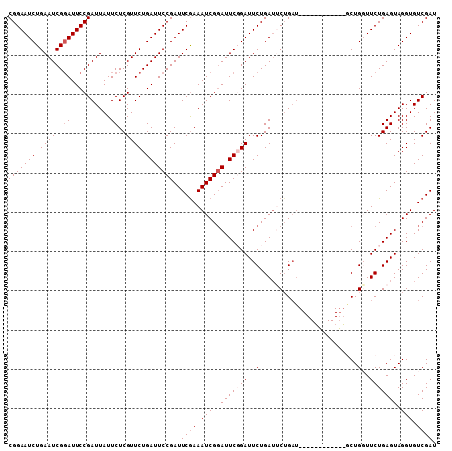
| Location | 19,854,725 – 19,854,827 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 95.13 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -22.34 |
| Energy contribution | -22.74 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19854725 102 + 20766785 -AUCUGAAUCAGAAUCCGAAUGCGAUUUCGAAUCGGAAUCAGAACGAGAAUAAUCGGAAUCCGAUUCAGAUUCCGCUUUCGGACCGUGUCAAUUGUUUGCAUU -.((((...)))).((((((.(((...((((((((((.((....(((......))))).)))))))).))...))).))))))..(((.(((....)))))). ( -30.90) >DroSec_CAF1 96725 102 + 1 -AUCAGCAUCAGAAUCCGAAUCCGAUUUCGAAUCGGAAUCAGAACGAGAAUAAUCGGAAUCCGAUUCAGAUUCCGCUUUCGGACCGUGUCAAUUGUUUGCAUU -....(((.(((((((.((.(((((((.....(((.........)))....))))))).)).))))).(((((((....))))....)))...))..)))... ( -27.30) >DroSim_CAF1 73607 102 + 1 -AUCAGAAUCAGAAUCCGAAUCCGAUUUCGAAUCGGAAUCAGAACGAGAAUAAUCGGAAUCCGAUUCAGAUUCCGCUUUCGGACCGUGUCAAUUGUUUGCAUU -....(((((.(((((.((.(((((((.....(((.........)))....))))))).)).))))).)))))......(((((.((....)).))))).... ( -26.60) >DroEre_CAF1 98523 103 + 1 ACUCAGACUCAGAAUCAGAAUCCGAUUUUGAAUCGGAAUCAGAACGAGAAUAAUCGGAAUCCGAUUCAGAUUCCGCAUUCGGACCGUGUCAAUUGUUUGCAUU .....(((.(.(((((.((.(((((((.....(((.........)))....))))))).)).)))))....((((....))))..).)))............. ( -27.20) >DroYak_CAF1 96554 103 + 1 AAGCAGCAUCAGAAUCAGAAUCCGAUUUCGAAUCGGAAUCAGAACGAGAAUAAUCGGAAUACGAUUCAGAUUCCGCUUUCGGACCGUGUCAAUUGUUUGCAUU ..((((((...(((((.((.((((((.....)))))).))....(((......)))......))))).(((((((....))))....)))...)).))))... ( -25.90) >consensus _AUCAGAAUCAGAAUCCGAAUCCGAUUUCGAAUCGGAAUCAGAACGAGAAUAAUCGGAAUCCGAUUCAGAUUCCGCUUUCGGACCGUGUCAAUUGUUUGCAUU ...........(((((.((.(((((((.....(((.........)))....))))))).)).)))))....((((....))))..(((.(((....)))))). (-22.34 = -22.74 + 0.40)

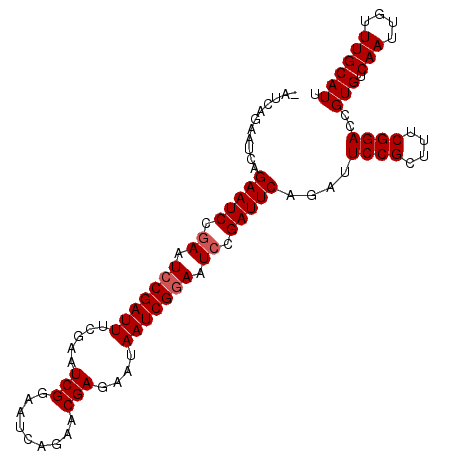
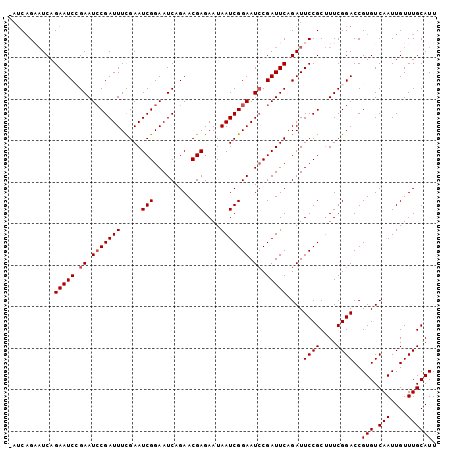
| Location | 19,854,725 – 19,854,827 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 95.13 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -24.84 |
| Energy contribution | -25.64 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19854725 102 - 20766785 AAUGCAAACAAUUGACACGGUCCGAAAGCGGAAUCUGAAUCGGAUUCCGAUUAUUCUCGUUCUGAUUCCGAUUCGAAAUCGCAUUCGGAUUCUGAUUCAGAU- ...........((((((..(((((((.((((..((.((((((((.((.((..........)).)).))))))))))..)))).)))))))..))..))))..- ( -33.70) >DroSec_CAF1 96725 102 - 1 AAUGCAAACAAUUGACACGGUCCGAAAGCGGAAUCUGAAUCGGAUUCCGAUUAUUCUCGUUCUGAUUCCGAUUCGAAAUCGGAUUCGGAUUCUGAUGCUGAU- ...(((..(....).((..(((((((..(((((((((...))))))))).................((((((.....)))))))))))))..)).)))....- ( -30.50) >DroSim_CAF1 73607 102 - 1 AAUGCAAACAAUUGACACGGUCCGAAAGCGGAAUCUGAAUCGGAUUCCGAUUAUUCUCGUUCUGAUUCCGAUUCGAAAUCGGAUUCGGAUUCUGAUUCUGAU- ...............((..(((.(((..(((((((((...)))))))))...........(((((.((((((.....)))))).)))))))).)))..))..- ( -31.00) >DroEre_CAF1 98523 103 - 1 AAUGCAAACAAUUGACACGGUCCGAAUGCGGAAUCUGAAUCGGAUUCCGAUUAUUCUCGUUCUGAUUCCGAUUCAAAAUCGGAUUCUGAUUCUGAGUCUGAGU .................(((..(((...(((((((((...))))))))).......)))..)))((((.((((((.((((((...)))))).)))))).)))) ( -30.10) >DroYak_CAF1 96554 103 - 1 AAUGCAAACAAUUGACACGGUCCGAAAGCGGAAUCUGAAUCGUAUUCCGAUUAUUCUCGUUCUGAUUCCGAUUCGAAAUCGGAUUCUGAUUCUGAUGCUGCUU .........................((((((.(((.((((((...(((((((....(((.((.......))..))))))))))...)))))).))).)))))) ( -25.80) >consensus AAUGCAAACAAUUGACACGGUCCGAAAGCGGAAUCUGAAUCGGAUUCCGAUUAUUCUCGUUCUGAUUCCGAUUCGAAAUCGGAUUCGGAUUCUGAUUCUGAU_ ...............((..(((((((..(((((((((...))))))))).................((((((.....)))))))))))))..))......... (-24.84 = -25.64 + 0.80)

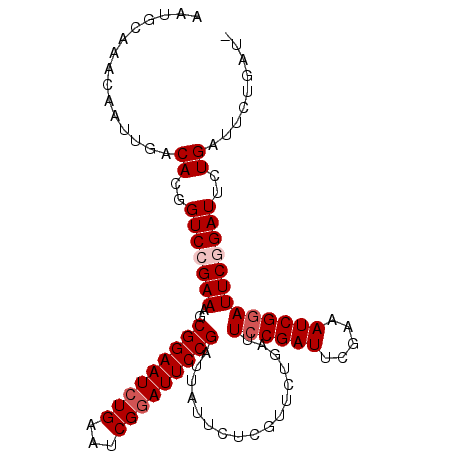
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:36 2006