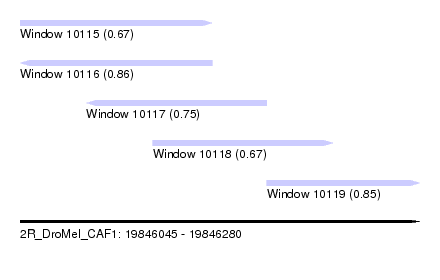
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,846,045 – 19,846,280 |
| Length | 235 |
| Max. P | 0.859673 |

| Location | 19,846,045 – 19,846,158 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 85.62 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -22.58 |
| Energy contribution | -24.00 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
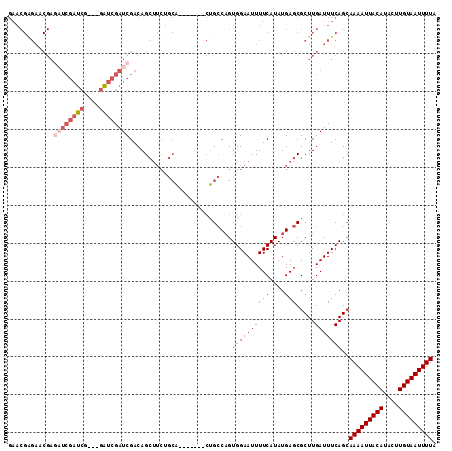
>2R_DroMel_CAF1 19846045 113 + 20766785 UGCUCUGAUUUCCAGGCAAAAGUCAAAGCCGGGCACAAAUUAAAAAUGCCCCACAAUUGCCCGCGAGUUUGAGAUCGGGAACGAGAACGAGAUCGAUCGGGCGAUCGAUCGAC (((.(((.....))))))...(((......(((((...........)))))..(.(((((((((((.((((...(((....)))...)))).)))..)))))))).)...))) ( -36.10) >DroSec_CAF1 88153 113 + 1 UGCUCUGAUUUCCAGGCAAAAGUCAAAGCCGGGCACAAAUUAAAAAUGCCCCACAAUUGCCCGGGAGCUUGAGAUCGGGAACGGGAACGAGAUCGAUCGAGCGAUCGAUCGAC ...(((((((((..(((....)))....(((((((......................)))))))......)))))))))..((....)).((((((((....))))))))... ( -38.15) >DroSim_CAF1 64384 113 + 1 UGCUCUGAUUUCCAGGCAAAAGUCAAAGCCGGGCACAAAUUAAAAAUGCCCCACAAUUGCCCGCGAGUUUGAGAUCGGGAACGAGAACGAGAUCGAUCAAGCGAUCGAUCGAC ..(((.(((((.(.(((((..((....)).(((((...........))))).....))))).).))))).))).(((....)))......((((((((....))))))))... ( -34.00) >DroEre_CAF1 89664 103 + 1 UGCUCUGAUUUCCAGGCAAAAGUCAAAGCCGGGCACAAAUUAAAAAUGCCCCACAAUUGGCCGCG------AGAUCGAGAACGAGAUCGAGAUCGAUCG----GUCGAUCGAC (((.(((.....))))))...(((......(((((...........)))))....((((((((((------(..((((........))))..)))..))----)))))).))) ( -32.10) >DroYak_CAF1 87598 93 + 1 UGCUCUGAUUACCAGGCAAAAGUCAAAGCCUGGCACAAAUUAAAAAUGCCCCACAAUUGCCGAUG------UGAUCGAGAACGAGAUCGA--------------UGGAUCGAC ..(((.((((((..(((((..((....))..((((...........))))......)))))...)------)))))))).....((((..--------------..))))... ( -21.90) >consensus UGCUCUGAUUUCCAGGCAAAAGUCAAAGCCGGGCACAAAUUAAAAAUGCCCCACAAUUGCCCGCGAG_UUGAGAUCGGGAACGAGAACGAGAUCGAUCG_GCGAUCGAUCGAC ..(((.((((((..(((..........)))(((((...........)))))...................)))))))))..((....)).((((((((....))))))))... (-22.58 = -24.00 + 1.42)


| Location | 19,846,045 – 19,846,158 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 85.62 |
| Mean single sequence MFE | -36.32 |
| Consensus MFE | -25.30 |
| Energy contribution | -28.04 |
| Covariance contribution | 2.74 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
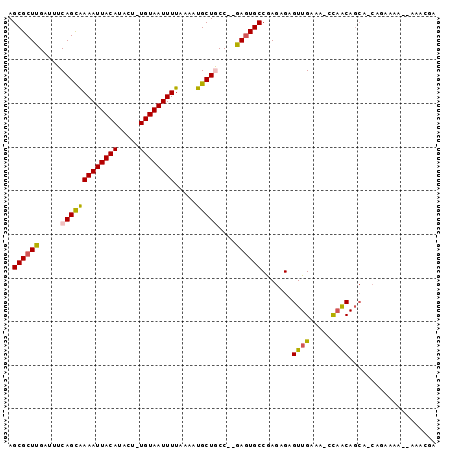
>2R_DroMel_CAF1 19846045 113 - 20766785 GUCGAUCGAUCGCCCGAUCGAUCUCGUUCUCGUUCCCGAUCUCAAACUCGCGGGCAAUUGUGGGGCAUUUUUAAUUUGUGCCCGGCUUUGACUUUUGCCUGGAAAUCAGAGCA ...((((((((....))))))))..(((((..(((((((........))).((((((..((.((((((.........)))))).))........))))))))))...))))). ( -41.00) >DroSec_CAF1 88153 113 - 1 GUCGAUCGAUCGCUCGAUCGAUCUCGUUCCCGUUCCCGAUCUCAAGCUCCCGGGCAAUUGUGGGGCAUUUUUAAUUUGUGCCCGGCUUUGACUUUUGCCUGGAAAUCAGAGCA ...((((((((....))))))))......................((((((((((((..((.((((((.........)))))).))........))))))))......)))). ( -40.60) >DroSim_CAF1 64384 113 - 1 GUCGAUCGAUCGCUUGAUCGAUCUCGUUCUCGUUCCCGAUCUCAAACUCGCGGGCAAUUGUGGGGCAUUUUUAAUUUGUGCCCGGCUUUGACUUUUGCCUGGAAAUCAGAGCA ...((((((((....))))))))..(((((..(((((((........))).((((((..((.((((((.........)))))).))........))))))))))...))))). ( -40.10) >DroEre_CAF1 89664 103 - 1 GUCGAUCGAC----CGAUCGAUCUCGAUCUCGUUCUCGAUCU------CGCGGCCAAUUGUGGGGCAUUUUUAAUUUGUGCCCGGCUUUGACUUUUGCCUGGAAAUCAGAGCA (((((((...----.))))))).........(((((.(((.(------(..((((((..((.((((((.........)))))).)).)))......)))..)).)))))))). ( -33.20) >DroYak_CAF1 87598 93 - 1 GUCGAUCCA--------------UCGAUCUCGUUCUCGAUCA------CAUCGGCAAUUGUGGGGCAUUUUUAAUUUGUGCCAGGCUUUGACUUUUGCCUGGUAAUCAGAGCA (((((....--------------)))))...(((((.(((((------((........)))))...............((((((((..........)))))))).))))))). ( -26.70) >consensus GUCGAUCGAUCGC_CGAUCGAUCUCGUUCUCGUUCCCGAUCUCAA_CUCGCGGGCAAUUGUGGGGCAUUUUUAAUUUGUGCCCGGCUUUGACUUUUGCCUGGAAAUCAGAGCA ...((((((((....))))))))..(((((((....)))...........(((((((..((.((((((.........)))))).))........))))))).......)))). (-25.30 = -28.04 + 2.74)

| Location | 19,846,084 – 19,846,190 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 87.98 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -24.43 |
| Energy contribution | -25.44 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19846084 106 - 20766785 CAUAUGAAAAUUCCACUGGCAG-------UGCAGAAGCUGUCGAUCGAUCGCCCGAUCGAUCUCGUUCUCGUUCCCGAUCUCAAACUCGCGGGCAAUUGUGGGGCAUUUUUAA ....((((((((((((.(((((-------(......))))))((((((((....))))))))........((((.(((........))).))))....)))))..))))))). ( -33.60) >DroSec_CAF1 88192 106 - 1 CAUAUGAAAAUUCCACUGGCAG-------UGCAGAAGCUGUCGAUCGAUCGCUCGAUCGAUCUCGUUCCCGUUCCCGAUCUCAAGCUCCCGGGCAAUUGUGGGGCAUUUUUAA ....(((((((......(((((-------(......))))))((((((((....))))))))..(((((((((((((............)))).)).)).)))))))))))). ( -33.10) >DroSim_CAF1 64423 106 - 1 CAUAUGAAAAUUCCACUGGCAG-------UGCAGAAGCUGUCGAUCGAUCGCUUGAUCGAUCUCGUUCUCGUUCCCGAUCUCAAACUCGCGGGCAAUUGUGGGGCAUUUUUAA ....((((((((((((.(((((-------(......))))))((((((((....))))))))........((((.(((........))).))))....)))))..))))))). ( -32.70) >DroEre_CAF1 89703 103 - 1 CAUAUGAAAAUUCCACUGGCAGAAGCCAGCGCAGAAGCUGUCGAUCGAC----CGAUCGAUCUCGAUCUCGUUCUCGAUCU------CGCGGCCAAUUGUGGGGCAUUUUUAA ....((((((((((.(((((....)))))(((((..(((((.((((((.----.(((((....)))))......)))))).------.)))))...)))))))).))))))). ( -36.10) >consensus CAUAUGAAAAUUCCACUGGCAG_______UGCAGAAGCUGUCGAUCGAUCGCUCGAUCGAUCUCGUUCUCGUUCCCGAUCUCAAACUCGCGGGCAAUUGUGGGGCAUUUUUAA ....((((((((((((.(((((...............)))))((((((((....))))))))........(..(.(((........))).)..)....)))))..))))))). (-24.43 = -25.44 + 1.00)

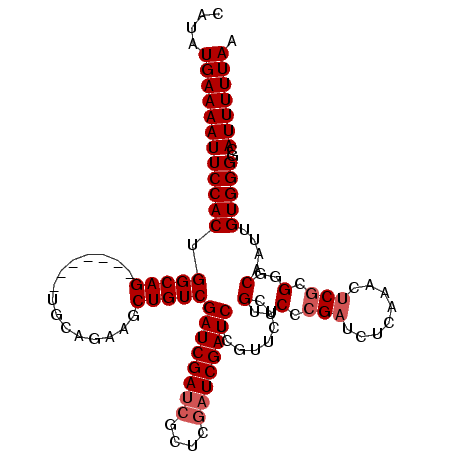
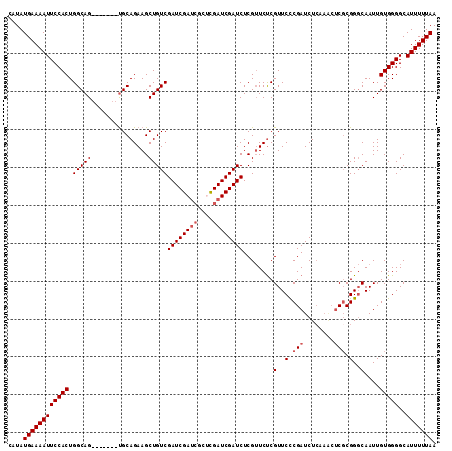
| Location | 19,846,123 – 19,846,229 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -15.04 |
| Energy contribution | -16.52 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
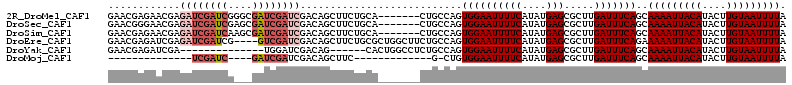
>2R_DroMel_CAF1 19846123 106 + 20766785 GAACGAGAACGAGAUCGAUCGGGCGAUCGAUCGACAGCUUCUGCA-------CUGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUA ....((((.((((.((((((((....))))))))..((....)).-------..(((((((((...))))).)).)).)))).))))...(((((((((....))))))))). ( -28.10) >DroSec_CAF1 88231 106 + 1 GAACGGGAACGAGAUCGAUCGAGCGAUCGAUCGACAGCUUCUGCA-------CUGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUA ...((....)).((((((((....))))))))....((....)).-------..(((((((((...))))).)).))(((.(....))))(((((((((....))))))))). ( -28.70) >DroSim_CAF1 64462 106 + 1 GAACGAGAACGAGAUCGAUCAAGCGAUCGAUCGACAGCUUCUGCA-------CUGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUA ....(((((...((((((((....))))))))......(((..(.-------......)..))))))))........(((.(....))))(((((((((....))))))))). ( -27.00) >DroEre_CAF1 89736 109 + 1 GAACGAGAUCGAGAUCGAUCG----GUCGAUCGACAGCUUCUGCGCUGGCUUCUGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGAAAAAUUACAUACUUGUAAUUUUA ....(((((((((.((((((.----...))))))..((((...(((((((....)))))))((....))....)))).)))))))))...(((((((((....))))))))). ( -40.40) >DroYak_CAF1 87670 93 + 1 GAACGAGAUCGA--------------UGGAUCGACAG------CACUGGCCUCUGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUA ....((((((((--------------((..(((....------(((((((....)))))))((....))...))).)).))))))))...(((((((((....))))))))). ( -26.70) >DroMoj_CAF1 167937 81 + 1 --------------UCGAUC----GAUCGAUCGACAGCUUC-------------G-CUGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUA --------------((((((----....))))))..(((.(-------------(-.((((((...)))))))))))(((.(....))))(((((((((....))))))))). ( -21.80) >consensus GAACGAGAACGAGAUCGAUCG___GAUCGAUCGACAGCUUCUGCA_______CUGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUA ............((((((((....))))))))...........................((((((((((....))).....)))))))..(((((((((....))))))))). (-15.04 = -16.52 + 1.47)
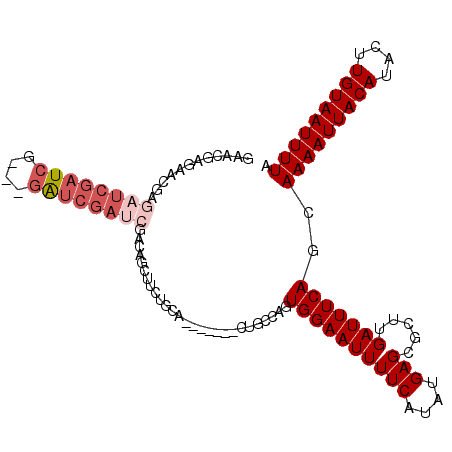
| Location | 19,846,190 – 19,846,280 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 85.21 |
| Mean single sequence MFE | -21.02 |
| Consensus MFE | -17.24 |
| Energy contribution | -17.13 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
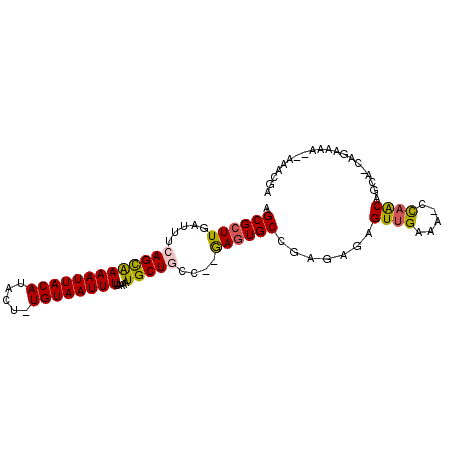
>2R_DroMel_CAF1 19846190 90 + 20766785 AGCGCUUGAUUUCAGCAAAAUUACAUACU-UGUAAUUUUAAAAUGCUGCC--GAGUGCCGAGAGAGUUGAAAAGCAACAGCA-CAAAA----AAACGC .(((((((....(((((((((((((....-)))))))).....))))).)--)))))).......((((.....))))....-.....----...... ( -22.20) >DroPse_CAF1 91601 96 + 1 AGCGCUUGAUUUCAGCAAAAUUACAUACU-UGUAAUUUUAAAAUGCUGCCAAGAGUGCCAAGAGAGUUGAAA-CCAACAGCAACAGAAAACUAAACGA .((((((.....(((((((((((((....-)))))))).....)))))....)))))).......((((...-.)))).................... ( -21.80) >DroWil_CAF1 207727 88 + 1 AGCGCUUGAUUUCAGUGAAAUUACAUACUUUGUAAUUUUAAAAUGCUACC--AACUGCCGAGAGAGUUGAAG-CUCACA-------AAAAAUAAAGGA .(((.(((.(..((.((((((((((.....))))))))))...))..).)--)).))).....((((....)-)))...-------............ ( -16.20) >DroYak_CAF1 87724 89 + 1 AGCGCUUGAUUUCAGCAAAAUUACAUACU-UGUAAUUUUAAAAUGCUGCC--GAGUGCCGAGAGAGUUGAGCAGCAGCAGCA-CAAA-----AAACGC .(((((.(....))))(((((((((....-)))))))))....((((((.--(...(((....).))....).)))))))).-....-----...... ( -23.10) >DroAna_CAF1 85209 92 + 1 AGCGCUUGAUUUCAGUAAAAUUACAUACU-UGUAAUUUUAAAAUGCUACC--GAGUGCCGAGAGAGUUGAA-AGCAACAGCA-CAGAAAAA-AAACGU .(((((((.(..((.((((((((((....-))))))))))...))..).)--)))))).......((((..-..))))....-........-...... ( -21.00) >DroPer_CAF1 92192 96 + 1 AGCGCUUGAUUUCAGCAAAAUUACAUACU-UGUAAUUUUAAAAUGCUGCCAAGAGUGCCAAGAGAGUUGAAA-CCAACAGCAACAGAAAACUAAACGA .((((((.....(((((((((((((....-)))))))).....)))))....)))))).......((((...-.)))).................... ( -21.80) >consensus AGCGCUUGAUUUCAGCAAAAUUACAUACU_UGUAAUUUUAAAAUGCUGCC__GAGUGCCGAGAGAGUUGAAA_CCAACAGCA_CAGAAAA__AAACGA .((((((.....(((((((((((((.....)))))))).....)))))....)))))).......((((.....)))).................... (-17.24 = -17.13 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:30 2006