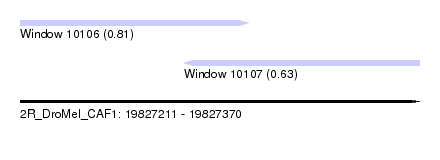
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,827,211 – 19,827,370 |
| Length | 159 |
| Max. P | 0.814647 |

| Location | 19,827,211 – 19,827,302 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 88.07 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -31.79 |
| Energy contribution | -31.16 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19827211 91 + 20766785 UCUGUGCAUGUCUUGGCCAUUGGAGGCUCCUAAGUCGGUCGGCUGGUCU--------AUUGGUGUGCUGGUCGGCUGGUCGGCUGGAGCGACUCCAGCC ..............(((...((((((((((..(((((..((((((..(.--------...........)..))))))..))))))))))..)))))))) ( -40.40) >DroSim_CAF1 57507 83 + 1 UCUGUGCAUGUCUUGGCCAUUGGAGGCUCCUAAGUCGGUCGGCUGGUCU--------AUUGGUGUGCUGGU--------CGGCUGGAGCGACUCCAGCC ..............(((...((((((((((..(((((..((((...((.--------...))...))))..--------))))))))))..)))))))) ( -33.40) >DroEre_CAF1 70952 99 + 1 GCUGUGCACGUCUUGGCCACUGGAGGCUCCUAAGUCGGUCGGCUGGCCUAUUGGACUAUUGGUGUGCUGGUCGGCUGGUCGGCUGGAGCGACUCCAGCC ..((.((........))))(((((((((((..(((((..(((((((((....(.((.......)).).)))))))))..))))))))))..)))))).. ( -46.60) >DroYak_CAF1 67664 91 + 1 UCGGUGCAUGUCUUGGCCACUGGAGGCUCCUAAGUCGGUCGGCUGGUCU--------AUUGGUGUGCUGGUGUGCUGGUCGGCUGGAGCGACUCCAGCC ..(((.((.....))))).(((((((((((......(((((((..((.(--------((..(....)..))).))..))))))))))))..)))))).. ( -39.50) >consensus UCUGUGCAUGUCUUGGCCACUGGAGGCUCCUAAGUCGGUCGGCUGGUCU________AUUGGUGUGCUGGUCGGCUGGUCGGCUGGAGCGACUCCAGCC .....((((..(..(((((((((.((((....))))..)))).)))))............)..)))).............((((((((...)))))))) (-31.79 = -31.16 + -0.63)

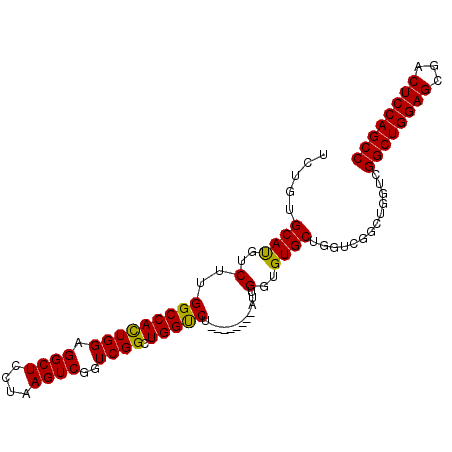
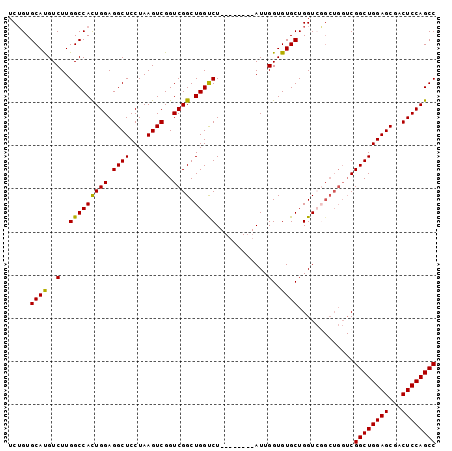
| Location | 19,827,276 – 19,827,370 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -28.06 |
| Energy contribution | -28.90 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19827276 94 - 20766785 GGGCCUGGUUAUAUCAUCGAGCACUCUUACGACGUGCCGCAGCCAAAUGGUCAACAGCUUGGUCGCUUGGCUGGAGUCGCUCCAGCCGACCAGC ....((((((........(.((((((....)).)))))((.(((((.((.....))..))))).))..((((((((...)))))))))))))). ( -37.00) >DroSim_CAF1 57570 88 - 1 GGGCCUGGUUAUAUCAUCGAGCACUCUUACGACGUGCCGCAGCCAAAUGGUCAACAGCUUGGUCGCUUGGCUGGAGUCGCUCCAGCCG------ .(((.(((.....((..((.((((((....)).))))))(((((((.((..(((....)))..)).)))))))))......)))))).------ ( -33.00) >DroEre_CAF1 71025 94 - 1 GGGCCUGGUUAUAUCAUCGAGCACUCUUACGACGUGCCGCAGCCAAAUGGUCAACAGCUUGGUCGCUUGGCUGGAGUCGCUCCAGCCGACCAGC ....((((((........(.((((((....)).)))))((.(((((.((.....))..))))).))..((((((((...)))))))))))))). ( -37.00) >DroYak_CAF1 67729 94 - 1 GGGCCUGGUUAUAUCAUCGAGCACUCUUACGACGUGCCGCAGCCAAAUGGUCAACAGCUUGGUCGCUUGGCUGGAGUCGCUCCAGCCGACCAGC ....((((((........(.((((((....)).)))))((.(((((.((.....))..))))).))..((((((((...)))))))))))))). ( -37.00) >DroAna_CAF1 65866 94 - 1 GGGCCUGGUUAUAUCAUCGAGCACUCUUACGACGUGCCGCAGCCAAAUGGUCAACAGCUUGGCCGCUUGGCUGGAGCCUCGAACGGCUAAGAGC .((((.((((.......((.((((((....)).))))))(((((((.(((((((....))))))).))))))).))))......))))...... ( -34.40) >consensus GGGCCUGGUUAUAUCAUCGAGCACUCUUACGACGUGCCGCAGCCAAAUGGUCAACAGCUUGGUCGCUUGGCUGGAGUCGCUCCAGCCGACCAGC .((((.((((.......((.((((((....)).))))))..(((....)))....)))).))))....((((((((...))))))))....... (-28.06 = -28.90 + 0.84)

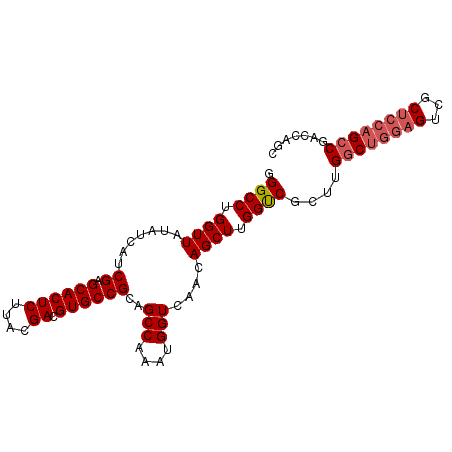
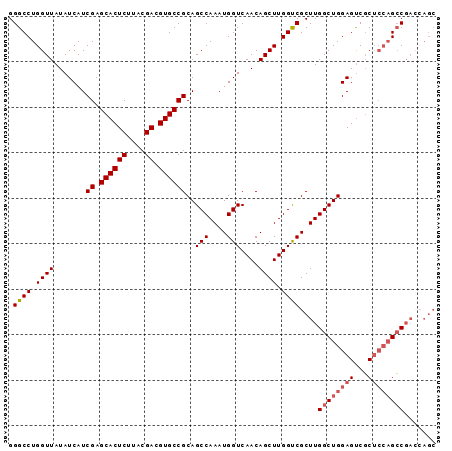
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:18 2006