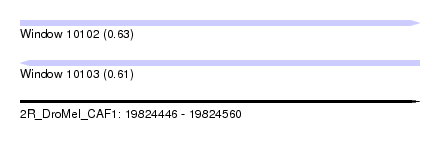
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,824,446 – 19,824,560 |
| Length | 114 |
| Max. P | 0.634657 |

| Location | 19,824,446 – 19,824,560 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -30.96 |
| Energy contribution | -31.20 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
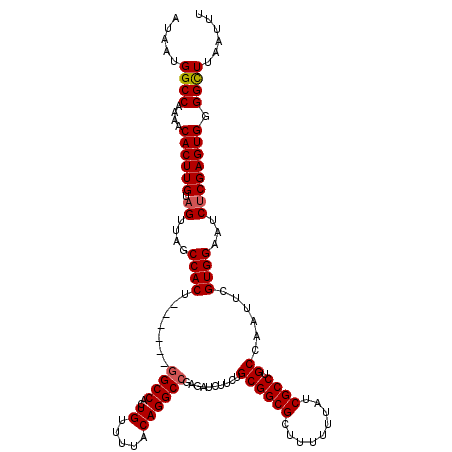
>2R_DroMel_CAF1 19824446 114 + 20766785 AUAAUGGCCAAAACACUUGUAGUUAGCCACU------GGCCACUGUUUUACAGGCCGAAAUCUUCUGCGGCGCUUUUUUAUCGCCUGCCAAUUCGUGGAAUCUCGAGUGGGGCUUAAUUU .....((((....((((((.((....(((((------((((..((.....))))))).........((((((.........)))).))......))))...)))))))).))))...... ( -33.80) >DroSec_CAF1 66707 114 + 1 AUAAUGGCCAAAACACUUGUAGUUAGCCACU------GGCCACUGUUUUACAGGCCGAGAUCUUCUGCGGCGCUUUUUUAUCGCCUGCCAAUUCGUGGAAUCUCGAGUGGGGCUUAAUUU .....((((....((((((.((....(((((------(((..(((.....)))((((((.....)).))))...............))))....))))...)))))))).))))...... ( -35.20) >DroSim_CAF1 54828 114 + 1 AUAAUGGCCAAAACACUUGUAGUUAGCCACU------GGCCACUGUUUUACAGGCCGAGAUCUUCUGCGGCGCUUUUUUAUCGCCUGCCAAUUCGUGGAAUCUCGAGUGGGGCUUAAUUU .....((((....((((((.((....(((((------(((..(((.....)))((((((.....)).))))...............))))....))))...)))))))).))))...... ( -35.20) >DroEre_CAF1 68233 113 + 1 CUAAUGGCCAAAACACUUGUAGUUAGCCACU------GGCCGCUGUUUUACAGGCCGAGAUCUUCUGCGGCGCUU-UUUAUCGCCUGCCAAUUCGUGGAAUCCCGAGUGGGGCUUAAUUU .....((((....((((((..((...(((((------((((...........))))).........((((((...-.....)))).))......)))).))..)))))).))))...... ( -34.00) >DroYak_CAF1 64763 120 + 1 AUAAUGGCCAAAACACUUGUAGUUGGCCACUGGUACUGGCCACUGUUUCACAGGCGGAGAUCUUGUGCGGCGCUUUUUUAUCGCCUGCCAAUUCGUGGAAUCUCGAGUGGGGUUUAAUUU .....((((....((((((((((.(((((.......))))))))).....(((((((.((....(((...)))....)).)))))))(((.....))).....)))))).))))...... ( -33.40) >consensus AUAAUGGCCAAAACACUUGUAGUUAGCCACU______GGCCACUGUUUUACAGGCCGAGAUCUUCUGCGGCGCUUUUUUAUCGCCUGCCAAUUCGUGGAAUCUCGAGUGGGGCUUAAUUU .....((((....((((((.((....((((.......((((..((.....))))))..........((((((.........)))).))......))))...)))))))).))))...... (-30.96 = -31.20 + 0.24)

| Location | 19,824,446 – 19,824,560 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -29.06 |
| Energy contribution | -28.98 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
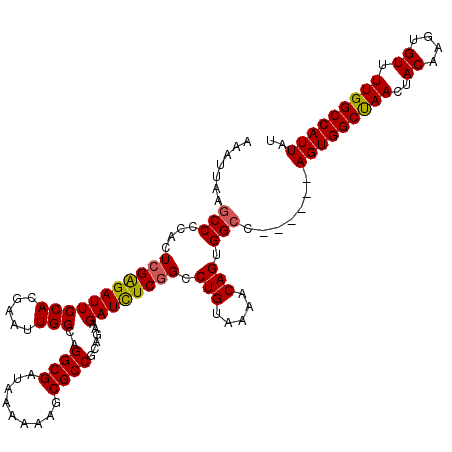
>2R_DroMel_CAF1 19824446 114 - 20766785 AAAUUAAGCCCCACUCGAGAUUCCACGAAUUGGCAGGCGAUAAAAAAGCGCCGCAGAAGAUUUCGGCCUGUAAAACAGUGGCC------AGUGGCUAACUACAAGUGUUUUGGCCAUUAU .......(((....(((........)))...))).((((.........))))............((((...........))))------(((((((((..((....)).))))))))).. ( -29.70) >DroSec_CAF1 66707 114 - 1 AAAUUAAGCCCCACUCGAGAUUCCACGAAUUGGCAGGCGAUAAAAAAGCGCCGCAGAAGAUCUCGGCCUGUAAAACAGUGGCC------AGUGGCUAACUACAAGUGUUUUGGCCAUUAU ................(((((((((.....)))..((((.........))))......))))))((((...........))))------(((((((((..((....)).))))))))).. ( -31.30) >DroSim_CAF1 54828 114 - 1 AAAUUAAGCCCCACUCGAGAUUCCACGAAUUGGCAGGCGAUAAAAAAGCGCCGCAGAAGAUCUCGGCCUGUAAAACAGUGGCC------AGUGGCUAACUACAAGUGUUUUGGCCAUUAU ................(((((((((.....)))..((((.........))))......))))))((((...........))))------(((((((((..((....)).))))))))).. ( -31.30) >DroEre_CAF1 68233 113 - 1 AAAUUAAGCCCCACUCGGGAUUCCACGAAUUGGCAGGCGAUAAA-AAGCGCCGCAGAAGAUCUCGGCCUGUAAAACAGCGGCC------AGUGGCUAACUACAAGUGUUUUGGCCAUUAG .......(((....(((((...)).)))...))).((((.....-...))))............((((.((......))))))------(((((((((..((....)).))))))))).. ( -32.90) >DroYak_CAF1 64763 120 - 1 AAAUUAAACCCCACUCGAGAUUCCACGAAUUGGCAGGCGAUAAAAAAGCGCCGCACAAGAUCUCCGCCUGUGAAACAGUGGCCAGUACCAGUGGCCAACUACAAGUGUUUUGGCCAUUAU ...........(((..((((((.......(((((.((((.........)))))).))))))))).....)))....((((((((((((..((((....))))..)))..))))))))).. ( -31.21) >consensus AAAUUAAGCCCCACUCGAGAUUCCACGAAUUGGCAGGCGAUAAAAAAGCGCCGCAGAAGAUCUCGGCCUGUAAAACAGUGGCC______AGUGGCUAACUACAAGUGUUUUGGCCAUUAU .......(((....(((((((((((.....)))..((((.........))))......)))))))).(((.....))).))).......(((((((((..((....)).))))))))).. (-29.06 = -28.98 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:15 2006