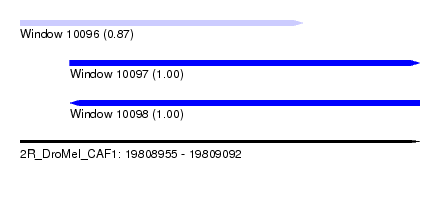
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,808,955 – 19,809,092 |
| Length | 137 |
| Max. P | 0.999944 |

| Location | 19,808,955 – 19,809,052 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.03 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.05 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19808955 97 + 20766785 GUUCGUUGG-----------------CUCU------CUUUUUUCGCGUUUCAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGGCCAGCAACCGCAACGCA (((.(((((-----------------(...------.......(((((..((((....))))......)))))((((((((((.........)))))))))))))))))))......... ( -23.30) >DroPse_CAF1 49434 100 + 1 GGCUAUUCC--------------UUUUUUU------UUCUUUUCGCGUUUCAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGACCAGCAACCGCAACGCA .........--------------.......------........(((((...(((..(((((........((.((((((((((.........)))))))))))))))))..)))))))). ( -22.90) >DroWil_CAF1 101569 84 + 1 ------------------------------------CUUUUUUCGCGUUUCAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGGCCAGCAACCGCAACGCA ------------------------------------........(((((...(((..(((((........((.((((((((((.........)))))))))))))))))..)))))))). ( -22.40) >DroMoj_CAF1 113128 98 + 1 ----AAUGG-----------------CUUGCCGACGUUU-UUUCGCGUUUCAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGGCCAGCAACCGCAACGCA ----..(((-----------------....)))......-....(((((...(((..(((((........((.((((((((((.........)))))))))))))))))..)))))))). ( -24.50) >DroAna_CAF1 47966 114 + 1 CGUCGUUGGCGUUGUCGUUGUAUUGUUGCU------UUUUUUCCGCGCUUCAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGGCCAGCAACCGCAACGCA ........(((((((.((((((((((.((.------........((((....))))..)).)))))...((..((((((((((.........))))))))))..))))))).))))))). ( -33.00) >DroPer_CAF1 49050 100 + 1 GGCUAUUCC--------------UUUUUUU------UUCUUUUCGCGUUUCAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGACCAGCAACCGCAACGCA .........--------------.......------........(((((...(((..(((((........((.((((((((((.........)))))))))))))))))..)))))))). ( -22.90) >consensus GG__AUUGG_________________UUUU______UUUUUUUCGCGUUUCAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGGCCAGCAACCGCAACGCA ............................................(((((...(((..(((((........((.((((((((((.........)))))))))))))))))..)))))))). (-22.10 = -22.05 + -0.05)

| Location | 19,808,972 – 19,809,092 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.56 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -34.63 |
| Energy contribution | -34.85 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.74 |
| SVM RNA-class probability | 0.999944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19808972 120 + 20766785 UUUCGCGUUUCAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGGCCAGCAACCGCAACGCAAUUUGGCAACACUUGUUGGCCAAUUGGCCAAAAUGUGAAG ....(((((...(((..(((((........((.((((((((((.........)))))))))))))))))..))))))))..........(((...((((((....))))))...)))... ( -36.60) >DroVir_CAF1 68366 120 + 1 UUUCGCGUUUCAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGGCCAGCAACCGCAACGCAAUUUGGCAACACUUGUUGGCCAAUUGGCAAAAACGUGAAA .(((((((((..(((..(((((........((.((((((((((.........)))))))))))))))))..)))...((...((((((((....))).)))))...))..))))))))). ( -38.30) >DroPse_CAF1 49454 120 + 1 UUUCGCGUUUCAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGACCAGCAACCGCAACGCAAUUUGGCAACACUUGUUGGCCAAUUGGCAAAAAUGUGAAG .(((((((((..(((..(((((........((.((((((((((.........)))))))))))))))))..)))...((...((((((((....))).)))))...))..))))))))). ( -37.00) >DroSec_CAF1 51459 120 + 1 UUUCGCGUUUCAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGGCCAGCAACCGCAACGCAAUUUGGCAACACUUGUUGGCCAAUUGGCCAAAAUAUGAAG .((((((((...(((..(((((........((.((((((((((.........)))))))))))))))))..))))))))....((....))....((((((....)))))).....))). ( -34.20) >DroSim_CAF1 30286 120 + 1 UUUCGCGUUUUAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGGCCAGCAACCGCAACGCAAUUUGGCAACACUUGUUGGCCAAUUGGCCAAAAUGUGAAG .((((((((((.(((..(((((........((.((((((((((.........)))))))))))))))))..)))...((...((((((((....))).)))))...)).)))))))))). ( -37.10) >DroAna_CAF1 48000 120 + 1 UUCCGCGCUUCAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGGCCAGCAACCGCAACGCAAUUUGGCAACACUUGUUGGCCAAUUGGCAAAAAUGUGAAG .......((((((((..(((((........((.((((((((((.........)))))))))))))))))..)))...((...((((((((....))).)))))...)).......))))) ( -32.50) >consensus UUUCGCGUUUCAGCGUCUGCUGCAAUUUAUGUGUAUAAAUGACAAAAGUACCGUUAUUUAUGGCCAGCAACCGCAACGCAAUUUGGCAACACUUGUUGGCCAAUUGGCAAAAAUGUGAAG .(((((((((..(((..(((((........((.((((((((((.........)))))))))))))))))..)))...((...((((((((....))).)))))...))..))))))))). (-34.63 = -34.85 + 0.22)

| Location | 19,808,972 – 19,809,092 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.56 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -30.64 |
| Energy contribution | -30.37 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19808972 120 - 20766785 CUUCACAUUUUGGCCAAUUGGCCAACAAGUGUUGCCAAAUUGCGUUGCGGUUGCUGGCCAUAAAUAACGGUACUUUUGUCAUUUAUACACAUAAAUUGCAGCAGACGCUGAAACGCGAAA ....(((((((((((....)))))..)))))).......((((((((((.((((((...((((((.((((.....)))).))))))...((.....)))))))).)))...))))))).. ( -34.20) >DroVir_CAF1 68366 120 - 1 UUUCACGUUUUUGCCAAUUGGCCAACAAGUGUUGCCAAAUUGCGUUGCGGUUGCUGGCCAUAAAUAACGGUACUUUUGUCAUUUAUACACAUAAAUUGCAGCAGACGCUGAAACGCGAAA .(((.((((((.((((((((((.(((....)))))).))))).(((((((((..((...((((((.((((.....)))).))))))...))..)))))))))....)).)))))).))). ( -35.10) >DroPse_CAF1 49454 120 - 1 CUUCACAUUUUUGCCAAUUGGCCAACAAGUGUUGCCAAAUUGCGUUGCGGUUGCUGGUCAUAAAUAACGGUACUUUUGUCAUUUAUACACAUAAAUUGCAGCAGACGCUGAAACGCGAAA .................(((((.(((....)))))))).((((((((((.((((((((.((((((.((((.....)))).)))))))).((.....)))))))).)))...))))))).. ( -31.40) >DroSec_CAF1 51459 120 - 1 CUUCAUAUUUUGGCCAAUUGGCCAACAAGUGUUGCCAAAUUGCGUUGCGGUUGCUGGCCAUAAAUAACGGUACUUUUGUCAUUUAUACACAUAAAUUGCAGCAGACGCUGAAACGCGAAA .(((.....((((((....))))))..((((((((......))(((((((((..((...((((((.((((.....)))).))))))...))..))))))))).)))))).......))). ( -33.00) >DroSim_CAF1 30286 120 - 1 CUUCACAUUUUGGCCAAUUGGCCAACAAGUGUUGCCAAAUUGCGUUGCGGUUGCUGGCCAUAAAUAACGGUACUUUUGUCAUUUAUACACAUAAAUUGCAGCAGACGCUAAAACGCGAAA ....(((((((((((....)))))..)))))).......((((((((((.((((((...((((((.((((.....)))).))))))...((.....)))))))).)))...))))))).. ( -34.20) >DroAna_CAF1 48000 120 - 1 CUUCACAUUUUUGCCAAUUGGCCAACAAGUGUUGCCAAAUUGCGUUGCGGUUGCUGGCCAUAAAUAACGGUACUUUUGUCAUUUAUACACAUAAAUUGCAGCAGACGCUGAAGCGCGGAA .............((..(((((.(((....))))))))...((((((((.((((((...((((((.((((.....)))).))))))...((.....)))))))).)))...))))))).. ( -30.90) >consensus CUUCACAUUUUGGCCAAUUGGCCAACAAGUGUUGCCAAAUUGCGUUGCGGUUGCUGGCCAUAAAUAACGGUACUUUUGUCAUUUAUACACAUAAAUUGCAGCAGACGCUGAAACGCGAAA .................(((((.(((....)))))))).((((((((((.((((((...((((((.((((.....)))).))))))...((.....)))))))).)))...))))))).. (-30.64 = -30.37 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:10 2006