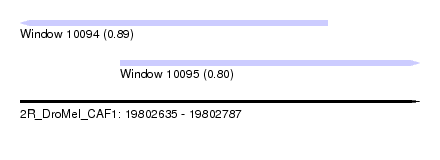
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,802,635 – 19,802,787 |
| Length | 152 |
| Max. P | 0.893150 |

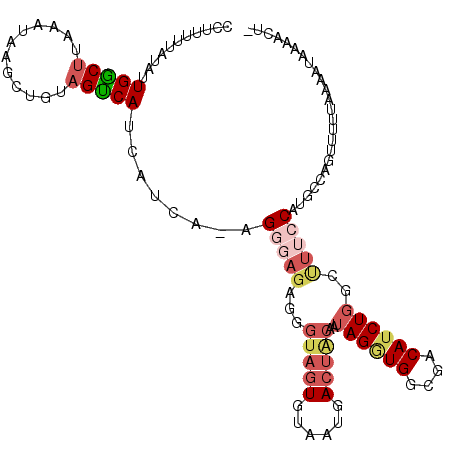
| Location | 19,802,635 – 19,802,752 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.76 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -3.93 |
| Energy contribution | -5.61 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.16 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.893150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19802635 117 - 20766785 UGUCCCCACCUAUUGUAGUCAUUACACUACCCUCUGUCUAUGAUGAUGACUACAGCUUAUUUAAGCCAAUAUAAAAAGGGGCAUAAAAAAUAUUAUCUAGUGAUCUCGUUAGGGAAA ((((((.......(((((((((((((..((.....))...)).)))))))))))((((....))))...........))))))....................((((....)))).. ( -27.20) >DroSec_CAF1 45069 111 - 1 UGUCGCCACCUAUUGUAGUCAUUACACUACC------CAAUGAUGAUGACUACAGCUUAUUUAAGUCAAUAUAAAAAGGGGCAUACAAAAUAUUAUAUUGCGAUCUCGUUAGGGAAA ....(((.(((..(((((((((((((.....------...)).)))))))))))((((....))))..........)))))).....................((((....)))).. ( -26.30) >DroSim_CAF1 23866 116 - 1 UGUCGCCACCUAUUGUAGUCAUUACACUACCCCCUCCCAGUGAUGAUGACUACACCUUAUUUAAGCCAAUAUAAAAAGGGGCAUACAAAAUAUUAUAUUGCGAUCUCGUUAGGGAA- ....(((......(((((((((((((((..........)))).)))))))))))((((.((((........))))))))))).....................((((....)))).- ( -28.60) >DroEre_CAF1 46059 98 - 1 UGUAGACAUCUACUGCC--------------CUCUCCCU---AUGAUGGCUACGAAUUAUGUAAGCCAAUAUAAUA--GAUCAUACAAAACAUUAUCUUACGAUCUUGCUUGGCAGA ..(((....)))(((((--------------.......(---(((.((((((((.....))).))))).))))..(--((((...................))))).....))))). ( -19.21) >DroYak_CAF1 42479 104 - 1 UGUGGCCAGCUACUGCACUCAUUACACUACGCUCUCCCU---AUGAUGGCUAGGAACUAUUUAGGCCAAUAUAAUA--G--------AAACAUUAUCUUACGAUCUUGUUGGGUGAG .((((....))))..................(((.((((---(((.(((((..((....))..))))).))))...--.--------.((((..(((....)))..))))))).))) ( -23.10) >consensus UGUCGCCACCUAUUGUAGUCAUUACACUACCCUCUCCCU_UGAUGAUGACUACAACUUAUUUAAGCCAAUAUAAAAAGGGGCAUACAAAAUAUUAUCUUGCGAUCUCGUUAGGGAAA ........(((..(((((((((((...................))))))))))).............................................(((....)))..)))... ( -3.93 = -5.61 + 1.68)

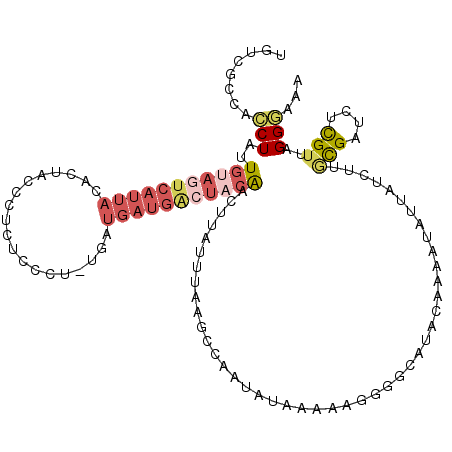
| Location | 19,802,673 – 19,802,787 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.49 |
| Mean single sequence MFE | -27.49 |
| Consensus MFE | -10.92 |
| Energy contribution | -12.86 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19802673 114 + 20766785 CCUUUUUAUAUUGGCUUAAAUAAGCUGUAGUCAUCAUCAUAGACAGAGGGUAGUGUAAUGACUACAAUAGGUGGGGACAUCUGGCUUUCCAUGCCAGUUUUUAAAAUAAAAUUU ...(((((.((((((......(((((((((((((...(((..((.....)).)))..))))))))).((((((....)))))))))).....))))))...)))))........ ( -33.50) >DroSec_CAF1 45107 108 + 1 CCUUUUUAUAUUGACUUAAAUAAGCUGUAGUCAUCAUCAUUG------GGUAGUGUAAUGACUACAAUAGGUGGCGACAUCUGGCUUUCCAUGCCGGUUUUUAAAAUAAAACUC .........................(((((((((...(((..------....)))..)))))))))..(((((....)))))(((.......)))((((((......)))))). ( -24.90) >DroSim_CAF1 23903 114 + 1 CCUUUUUAUAUUGGCUUAAAUAAGGUGUAGUCAUCAUCACUGGGAGGGGGUAGUGUAAUGACUACAAUAGGUGGCGACAUCUGGCUUUCCAUGCCGGUUUCUAAAAUAAAACUC ...(((((.((((((......(((((((((((((...(((((........)))))..))))))))..((((((....)))))))))))....))))))...)))))........ ( -31.40) >DroEre_CAF1 46097 94 + 1 C--UAUUAUAUUGGCUUACAUAAUUCGUAGCCAUCAU---AGGGAGAG--------------GGCAGUAGAUGUCUACAUCUGGCCUUCCAUACCAGUUAUUAAAAUAAAACU- .--...(((..(((((.((.......)))))))..))---)((..(((--------------(((..((((((....))))))))))))....))..................- ( -24.20) >DroYak_CAF1 42509 107 + 1 C--UAUUAUAUUGGCCUAAAUAGUUCCUAGCCAUCAU---AGGGAGAGCGUAGUGUAAUGAGUGCAGUAGCUGGCCACAUCUUGCUUUCCAUUCCAGUUAUUUAAAUUAAAC-- .--..((((..((((..............))))..))---))((((((((..((((..(.(((......))).)..))))..))))))))......................-- ( -23.44) >consensus CCUUUUUAUAUUGGCUUAAAUAAGCUGUAGUCAUCAUCA_AGGGAGAGGGUAGUGUAAUGACUACAAUAGGUGGCGACAUCUGGCUUUCCAUGCCAGUUUUUAAAAUAAAACU_ ...........(((((............)))))........(((((...(((((......)))))..((((((....))))))..)))))........................ (-10.92 = -12.86 + 1.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:07 2006