| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,793,231 – 19,793,323 |
| Length | 92 |
| Max. P | 0.948724 |

| Location | 19,793,231 – 19,793,323 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 71.97 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -23.04 |
| Energy contribution | -23.64 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
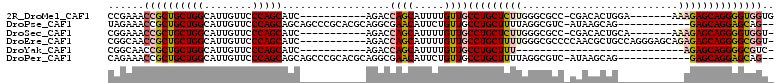
>2R_DroMel_CAF1 19793231 92 + 20766785 CCGAAACCGCUGCUGGCAUUGUUCCCAGCAUC-----------AGACCAGCAUUUUGUUGCCUGCUCUUGGGCGCC-CGACACUGGA-------AAAGAGCAGGGGUGGUG .....((((((((((((..(((.....)))..-----------.).))))).........(((((((((.....((-.......)).-------.))))))))))))))). ( -33.40) >DroPse_CAF1 33350 96 + 1 UAGAAACCGCUGCUGGCAUUGUUCCCAGCAGCAGCCCGCACGCAGGCGAACAUUCUGUUGCCUGCUUUUAGGCGUC-AUAAGCAG------------GAGCAGGAGCAG-- ........((((((((........)))))))).(((.(....).))).......(((((.((((((((...((...-....)).)------------))))))))))))-- ( -38.80) >DroSec_CAF1 35718 91 + 1 CGGAAACCGCUGCUGGCAUUGUUCCCAGCAUC-----------AGACCAGCAUUUUGUUGCCUGCUCUUGGGCGCC-CGACACUGCA-------AAAGAGCAGGGGUGGU- .....((((((((((((..(((.....)))..-----------.).))))).........(((((((((..(((..-......))).-------.)))))))))))))))- ( -36.10) >DroEre_CAF1 35202 99 + 1 CGGCAACCGCUGCUGGCAUUGUUCCCAGCAUC-----------AGACCAGCAUUUUGUUGCCUGCUUUUGGGCGCCCCAACGCUGCCAGGGAGCAGAGAGCAGGGGCGGU- .....((((((((((((..(((.....)))..-----------.).)))))..((((((..(((((((..(((((......)).)))..)))))))..))))))))))))- ( -40.00) >DroYak_CAF1 32884 71 + 1 CGGCAACCGCUGCUGGCAUUGUUCCCAGCAUC-----------AGACCAGCAUUUUGUUGCCUGCUUU----------------------------AGAGCAGGGGCGUC- .(....)((((((((((..(((.....)))..-----------.).))))).........((((((..----------------------------..))))))))))..- ( -24.10) >DroPer_CAF1 32970 96 + 1 CAGAAACCGCUGCUGGCAUUGUUCCCAGCAGCAGCCCGCACGCAGGCGAACAUUCUGUUGCCUGCUUUUAGGCGUC-AUAAGCAG------------GAGCAGGAGCAG-- ........((((((((........)))))))).(((.(....).))).......(((((.((((((((...((...-....)).)------------))))))))))))-- ( -38.80) >consensus CGGAAACCGCUGCUGGCAUUGUUCCCAGCAUC___________AGACCAGCAUUUUGUUGCCUGCUUUUGGGCGCC_CGACGCUG___________AGAGCAGGGGCGGU_ ......((((((((((........)))))..................((((.....))))(((((((((..........................)))))))))))))).. (-23.04 = -23.64 + 0.60)
| Location | 19,793,231 – 19,793,323 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.97 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
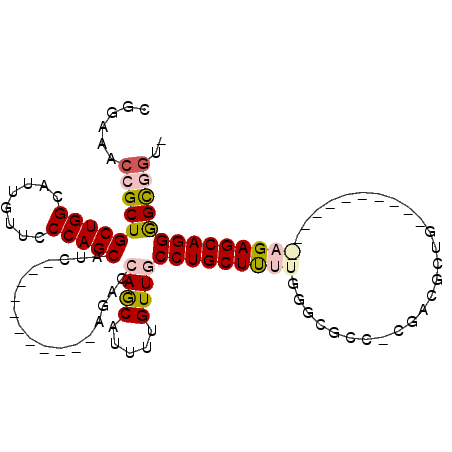
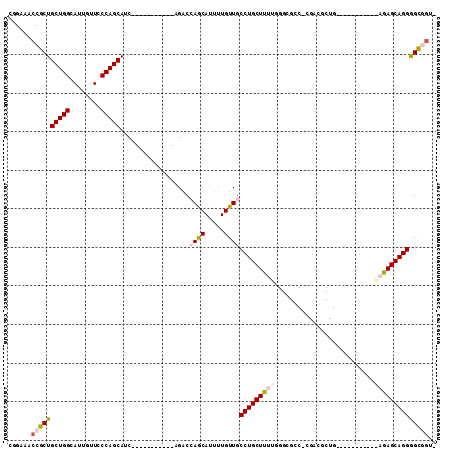
>2R_DroMel_CAF1 19793231 92 - 20766785 CACCACCCCUGCUCUUU-------UCCAGUGUCG-GGCGCCCAAGAGCAGGCAACAAAAUGCUGGUCU-----------GAUGCUGGGAACAAUGCCAGCAGCGGUUUCGG ..(((((.(((((..((-------((((((((((-(((((......)).((((......)))).))))-----------))))))))))).......))))).)))...)) ( -34.30) >DroPse_CAF1 33350 96 - 1 --CUGCUCCUGCUC------------CUGCUUAU-GACGCCUAAAAGCAGGCAACAGAAUGUUCGCCUGCGUGCGGGCUGCUGCUGGGAACAAUGCCAGCAGCGGUUUCUA --((((.(..((..------------..))....-...........(((((((((.....))).))))))).))))(((((((((((........)))))))))))..... ( -40.10) >DroSec_CAF1 35718 91 - 1 -ACCACCCCUGCUCUUU-------UGCAGUGUCG-GGCGCCCAAGAGCAGGCAACAAAAUGCUGGUCU-----------GAUGCUGGGAACAAUGCCAGCAGCGGUUUCCG -......(((((((((.-------.((.((....-.))))..)))))))))............((.((-----------(.((((((........)))))).)))...)). ( -31.10) >DroEre_CAF1 35202 99 - 1 -ACCGCCCCUGCUCUCUGCUCCCUGGCAGCGUUGGGGCGCCCAAAAGCAGGCAACAAAAUGCUGGUCU-----------GAUGCUGGGAACAAUGCCAGCAGCGGUUGCCG -..((((((.((...(((((....))))).)).))))))..........((((((....(((((((..-----------.....((....))..)))))))...)))))). ( -41.40) >DroYak_CAF1 32884 71 - 1 -GACGCCCCUGCUCU----------------------------AAAGCAGGCAACAAAAUGCUGGUCU-----------GAUGCUGGGAACAAUGCCAGCAGCGGUUGCCG -........((((..----------------------------..))))((((((....(((((((..-----------.....((....))..)))))))...)))))). ( -24.80) >DroPer_CAF1 32970 96 - 1 --CUGCUCCUGCUC------------CUGCUUAU-GACGCCUAAAAGCAGGCAACAGAAUGUUCGCCUGCGUGCGGGCUGCUGCUGGGAACAAUGCCAGCAGCGGUUUCUG --((((.(..((..------------..))....-...........(((((((((.....))).))))))).))))(((((((((((........)))))))))))..... ( -40.10) >consensus _ACCGCCCCUGCUCU___________CAGCGUAG_GGCGCCCAAAAGCAGGCAACAAAAUGCUGGUCU___________GAUGCUGGGAACAAUGCCAGCAGCGGUUUCCG ..((((.((((((................................))))))..............................((((((........))))))))))...... (-18.56 = -18.83 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:02 2006