
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
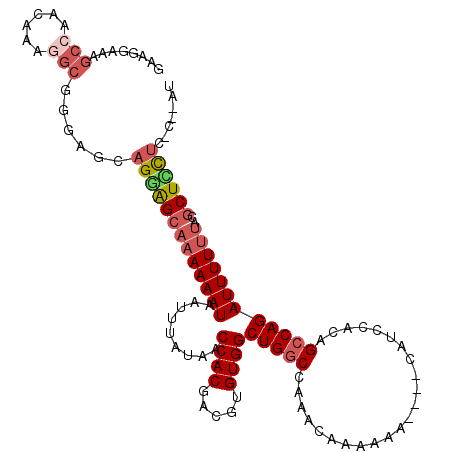
| Location | 19,752,773 – 19,752,882 |
| Length | 109 |
| Max. P | 0.805409 |

| Location | 19,752,773 – 19,752,882 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -17.42 |
| Energy contribution | -18.53 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
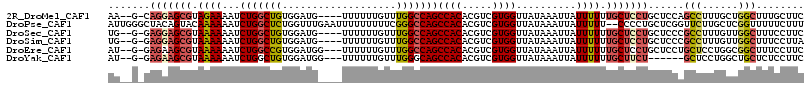
>2R_DroMel_CAF1 19752773 109 + 20766785 GAAGCAAAGCCAGCAAAGGCUGGAGCAGGAGCAAAAAAUAAUUUAUAACCACGACGUGUGGCUGGCCAAACAAAAAA----CAUCCACAGCCAGAUUUUCUACGCUCCUG-C--UU .........(((((....)))))(((((((((................((((.....))))(((((...........----........))))).........)))))))-)--). ( -36.31) >DroPse_CAF1 13290 114 + 1 AAAGAAAAACCGAGCAAGAACCGAGCAGGGG--AAAAAUAAUUUAUAACCACGACGUGUGGCUGGCCCGAAAAAAAAAUUCAAACCAGAGCCAGAUUUUUUGUACUGUAGCCCAAU ........................(((((.(--((((((.........((((.....))))(((((.(...................).)))))))))))).).))))........ ( -18.21) >DroSec_CAF1 5589 109 + 1 GAAGGAAAGCCAACAAAGGCGGGAGCAGGAGCAAAAAAUAAUUUAUAACCACGACGUGUGGCUGGCCAAACAAAAAA----CAUCCACAGCCAGAUUUUUUACGCUCCUC-C--CA ........(((......)))((((...((((((((((((.........((((.....))))(((((...........----........))))))))))))..)))))))-)--). ( -31.51) >DroSim_CAF1 8610 109 + 1 UAAGGAAAGCCAACAAAGGCGGGAGCAGGAGCAAAAAAUAAUUUAUAACCACGACGUGUGGCUGGCCAAACAAAAAA----CAUCCACAGCCAGAUUUUUUACGCUCCUC-C--CA ........(((......)))((((...((((((((((((.........((((.....))))(((((...........----........))))))))))))..)))))))-)--). ( -31.51) >DroEre_CAF1 5733 110 + 1 GAAGGAAAGCCGCCAGGAGCAGGAGCAGGAGCAAAAAAUAAUUUAUAACCACGACGUGUGGCUGGCCAAACAAAAAA---CCAUCCACGGCCAGAUUUUUUACGCUUCUC-C--AU ...(((..((..((.......)).)).((((((((((((.........((((.....))))((((((..........---........)))))))))))))..)))))))-)--.. ( -27.87) >DroYak_CAF1 8541 104 + 1 GAAGGAGAGCAGCCAGGAGC------AGAAGCAAAAAAUAAUUUAUAACCACGACGUGUGGCUGCCCAAACAAAAAA---CCAUCCACAGCCAGAUUUUUUACGCUUCUC-C--AU ...((((((((((((...((------....)).................(((...))))))))))............---........(((............)))))))-)--.. ( -21.60) >consensus GAAGGAAAGCCAACAAAGGCGGGAGCAGGAGCAAAAAAUAAUUUAUAACCACGACGUGUGGCUGGCCAAACAAAAAA____CAUCCACAGCCAGAUUUUUUACGCUCCUC_C__AU ........(((......)))......(((((((((((((.........((((.....))))(((((.......................))))))))))))..))))))....... (-17.42 = -18.53 + 1.11)

| Location | 19,752,773 – 19,752,882 |
|---|---|
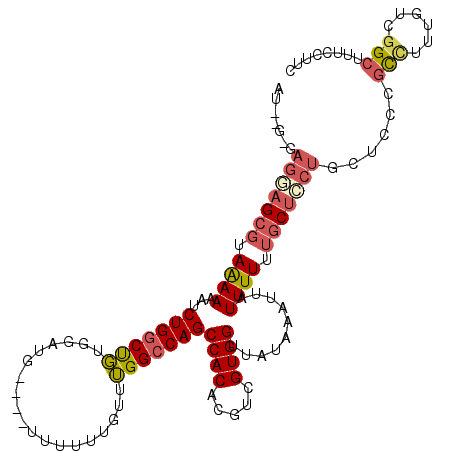
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -20.61 |
| Energy contribution | -21.31 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19752773 109 - 20766785 AA--G-CAGGAGCGUAGAAAAUCUGGCUGUGGAUG----UUUUUUGUUUGGCCAGCCACACGUCGUGGUUAUAAAUUAUUUUUUGCUCCUGCUCCAGCCUUUGCUGGCUUUGCUUC .(--(-((((((((.(((((..(((((((..((..----...))..)..))))))((((.....))))..........)))))))))))))))(((((....)))))......... ( -39.30) >DroPse_CAF1 13290 114 - 1 AUUGGGCUACAGUACAAAAAAUCUGGCUCUGGUUUGAAUUUUUUUUUCGGGCCAGCCACACGUCGUGGUUAUAAAUUAUUUUU--CCCCUGCUCGGUUCUUGCUCGGUUUUUCUUU (((((((...((.......((((.(((.((((((((((.......))))))))))((((.....))))...............--.....))).)))))).)))))))........ ( -24.71) >DroSec_CAF1 5589 109 - 1 UG--G-GAGGAGCGUAAAAAAUCUGGCUGUGGAUG----UUUUUUGUUUGGCCAGCCACACGUCGUGGUUAUAAAUUAUUUUUUGCUCCUGCUCCCGCCUUUGUUGGCUUUCCUUC .(--(-(((.((.((((((((((((((((..((..----...))..)..))))))((((.....)))).........)))))))))..)).)))))(((......)))........ ( -36.40) >DroSim_CAF1 8610 109 - 1 UG--G-GAGGAGCGUAAAAAAUCUGGCUGUGGAUG----UUUUUUGUUUGGCCAGCCACACGUCGUGGUUAUAAAUUAUUUUUUGCUCCUGCUCCCGCCUUUGUUGGCUUUCCUUA .(--(-(((.((.((((((((((((((((..((..----...))..)..))))))((((.....)))).........)))))))))..)).)))))(((......)))........ ( -36.40) >DroEre_CAF1 5733 110 - 1 AU--G-GAGAAGCGUAAAAAAUCUGGCCGUGGAUGG---UUUUUUGUUUGGCCAGCCACACGUCGUGGUUAUAAAUUAUUUUUUGCUCCUGCUCCUGCUCCUGGCGGCUUUCCUUC ..--(-((((.((((((((((((((((((..((..(---....)..)))))))))((((.....)))).........))))))))).((.((....))....))..)))))))... ( -31.00) >DroYak_CAF1 8541 104 - 1 AU--G-GAGAAGCGUAAAAAAUCUGGCUGUGGAUGG---UUUUUUGUUUGGGCAGCCACACGUCGUGGUUAUAAAUUAUUUUUUGCUUCU------GCUCCUGGCUGCUCUCCUUC ..--(-((((.(((((((((((.((((((((((..(---....)..)))..)))))))(((...)))..........)))))))))....------((.....)).)))))))... ( -28.80) >consensus AU__G_GAGGAGCGUAAAAAAUCUGGCUGUGGAUG____UUUUUUGUUUGGCCAGCCACACGUCGUGGUUAUAAAUUAUUUUUUGCUCCUGCUCCCGCCUUUGUCGGCUUUCCUUC .......(((((((.((((...(((((((...................)))))))((((.....))))..........)))).)))))))......(((......)))........ (-20.61 = -21.31 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:49 2006