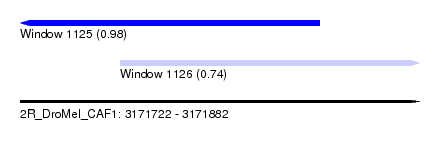
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,171,722 – 3,171,882 |
| Length | 160 |
| Max. P | 0.984631 |

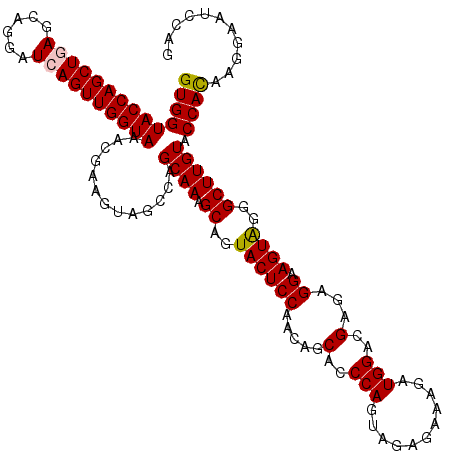
| Location | 3,171,722 – 3,171,842 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -28.38 |
| Energy contribution | -28.26 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3171722 120 - 20766785 UCUUCCUCUACUGGGUGCUGUUGGAGUACUGCUUUGCUGGCUAUUUCAUUUACCAACUCAUCCUGCUCAGCUGGUACCACACCAUUAUUCUGUUCCUGCAGCGCUUCAUCGAGGACCAUG ...(((((....(((((((((.((((.........((((((.......................)).))))((((.....))))........)))).)))))))))....)))))..... ( -33.70) >DroSec_CAF1 61289 120 - 1 UCUUUCUCUACUGGGUGCUGUUGGAGUACUGCUUUGCUGGCUACUUCGUUUACCAACUGAUCCUGCUCAGCUGGUACCACACCAUUAUUCUGUUCCUGCAGCGCUUCAUCGAGGACCAUG ...(((((....(((((((((.((((....(((.....)))......((.(((((.((((......)))).)))))..))............)))).)))))))))....)))))..... ( -33.70) >DroSim_CAF1 63127 120 - 1 UCUUCCUCUACUGGGUGCUGUUGGAGUACUGCUUUGCUGGCUACUUCGUUUACCAACUGAUCCUGCUCAGCUGGUACCACACAAUUAUUCUGUUCCUGCAGCGCUUCAUCGAGGACCAUG ...(((((....(((((((((.((((....(((.....)))......((.(((((.((((......)))).)))))..))............)))).)))))))))....)))))..... ( -36.60) >DroEre_CAF1 60237 120 - 1 UCUUUCUCUACUGGGUGCUGAUGGAGUACUGCUUUGCUGCCUACUUCGUUUACCAACUUAUCCUGCUCAGCUGGUACCACACCAUUAUUUUGUUCCUGCAGCGCCUUAUCGAGGACCAUG ...(((((....((((((((((((((((..((......)).)))))))).(((((.((..........)).)))))......................))))))))....)))))..... ( -30.70) >DroYak_CAF1 60992 120 - 1 UCUUUCUUUACUGGCUGCUGUUGGAGUACUGCUUUGCAGGGUACUUCGUUUACCAACUGAUCCUGCUCAGCUGGUACCACACCAUUAUCCUGUUCCUCCAGCACUUUAUCGCGGACCAUG ...........(((((((...((((((.(((....((((((((....((.(((((.((((......)))).)))))..)).....)))))))).....))).))))))..)))).))).. ( -35.50) >consensus UCUUUCUCUACUGGGUGCUGUUGGAGUACUGCUUUGCUGGCUACUUCGUUUACCAACUGAUCCUGCUCAGCUGGUACCACACCAUUAUUCUGUUCCUGCAGCGCUUCAUCGAGGACCAUG ...(((((....((((((((..(((((((((((..((.((.....................)).))..))).))))).(((.........))))))..))))))))....)))))..... (-28.38 = -28.26 + -0.12)

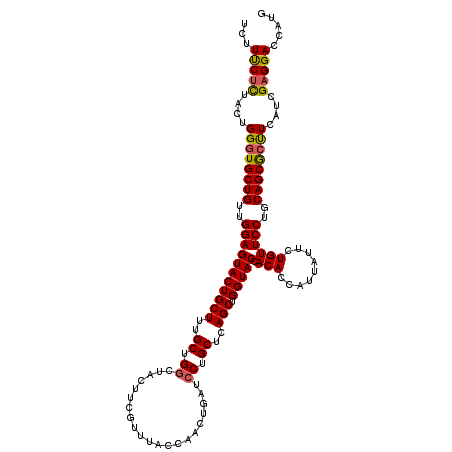
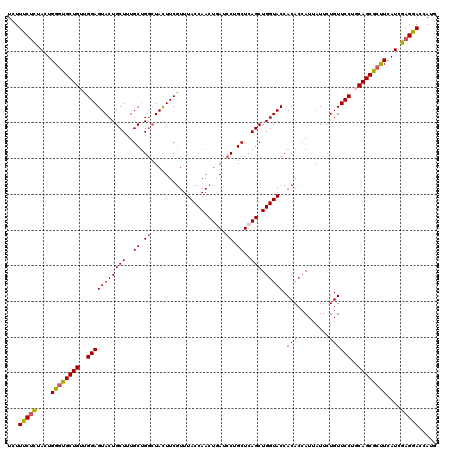
| Location | 3,171,762 – 3,171,882 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -30.00 |
| Energy contribution | -30.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3171762 120 + 20766785 GUGGUACCAGCUGAGCAGGAUGAGUUGGUAAAUGAAAUAGCCAGCAAAGCAGUACUCCAACAGCACCCAGUAGAGGAAGAUGGACGAGAGGAAGUAGGGCUUGUACCACAAGGAAUCCAG (((((((.((((..((.......((((((..........)))))).........(((...((....((......))....))...))).....))..)))).)))))))........... ( -30.90) >DroSec_CAF1 61329 120 + 1 GUGGUACCAGCUGAGCAGGAUCAGUUGGUAAACGAAGUAGCCAGCAAAGCAGUACUCCAACAGCACCCAGUAGAGAAAGAUGGAGGAGAGGAAGUAGGGCUUGUACCACAAGGAAUCCAG ((((((((((((((......)))))))))..............((((.((..((((((.....(..(((...........)))..)...)).))))..)))))))))))........... ( -34.30) >DroSim_CAF1 63167 120 + 1 GUGGUACCAGCUGAGCAGGAUCAGUUGGUAAACGAAGUAGCCAGCAAAGCAGUACUCCAACAGCACCCAGUAGAGGAAGAUGGAGGAGAGGAAGUAGGGCUUGUACCACAAGGAAUCCAG (((((((.((((..((.......((((((..........)))))).........((((..((....((......))....))..)))).....))..)))).)))))))........... ( -35.40) >DroEre_CAF1 60277 120 + 1 GUGGUACCAGCUGAGCAGGAUAAGUUGGUAAACGAAGUAGGCAGCAAAGCAGUACUCCAUCAGCACCCAGUAGAGAAAGAUGGACGAGAGGAAGUGGGGCUUGUGCCAUAAGGAAUCCAG .(((((((((((..........)))))))).........((((...((((..(((((((((.................))))))(....)...)))..)))).)))).........))). ( -31.53) >DroYak_CAF1 61032 120 + 1 GUGGUACCAGCUGAGCAGGAUCAGUUGGUAAACGAAGUACCCUGCAAAGCAGUACUCCAACAGCAGCCAGUAAAGAAAGAUGGACGAGAGGAAGUAGGGCUUGUUCCAUAAGGAUUCCAG ((((((((((((((......))))))))))(((((....((((((...((.((......)).)).((((...........))).)........)))))).)))))))))........... ( -32.00) >consensus GUGGUACCAGCUGAGCAGGAUCAGUUGGUAAACGAAGUAGCCAGCAAAGCAGUACUCCAACAGCACCCAGUAGAGAAAGAUGGACGAGAGGAAGUAGGGCUUGUACCACAAGGAAUCCAG ((((((((((((((......)))))))))).............((((.((..((((((.....(..(((...........)))..)...)).))))..)))))).))))........... (-30.00 = -30.00 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:12 2006