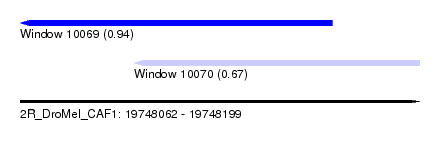
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,748,062 – 19,748,199 |
| Length | 137 |
| Max. P | 0.938840 |

| Location | 19,748,062 – 19,748,169 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.56 |
| Mean single sequence MFE | -40.38 |
| Consensus MFE | -27.83 |
| Energy contribution | -28.70 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.938840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
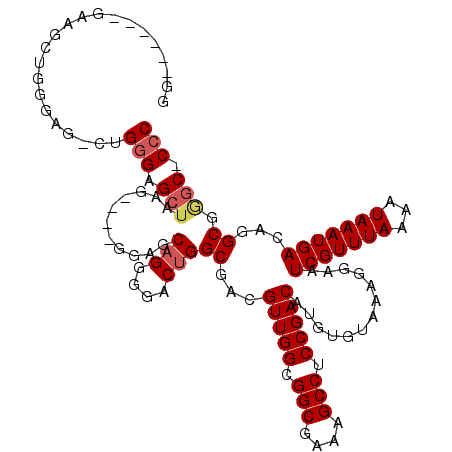
>2R_DroMel_CAF1 19748062 107 - 20766785 G-------GAAGCUGGGAG-CUGGGAGCUAAG----GGAGCAGGGGACUGGCGACGUUGGCGGCGAAAGCCUCCGACAUGUGUAAAGGAAUCGUUUAAAAUAAAUGACAGGCGGAC-CCC .-------...(((...((-(.....)))...----..))).((((.(((.(...(((((.(((....))).))))).............(((((((...)))))))..).))).)-))) ( -36.50) >DroSec_CAF1 851 114 - 1 GGGAGCUGGAAGCUGGGAG-CUGGGAGCUAAG----GGAGCAGGGGACUGGCGACGUUGGCGGCGAAAGCCUCCGACAUGUGUAAAGGAAUCGUUUAAAAUAAAUGACAGGCCGGC-CCC (((.(((((...(((....-...((((((((.----(..((((....)).))..).)))))(((....))))))................(((((((...)))))))))).)))))-))) ( -45.10) >DroSim_CAF1 3356 100 - 1 GG--------------GAG-CUGGGAGCUAAG----GGAGCAGGGGACCGGCGACGUUGGCGGCGAAAGCCUCCGACAUGUGUAAAGGAAUCGUUUAAAAUAAAUGACAGGCGAGC-CCC ((--------------(.(-((.(..(((((.----(..((.(....)..))..).)))))(((....)))(((.((....))...))).(((((((...)))))))....).)))-))) ( -35.80) >DroYak_CAF1 3459 117 - 1 GU-GGCAAGUGGCAGG-GGGUAG-GGGCAUGGGGCAGGGGCAGGGGACUGGCGACGUUGGCGGCGAAAGCCUCCGACAUGUGUAAAGGAAUCGUUUAAAAUAAAUGACAGGCGGGCCCCC ..-.((.....)).((-((((..-..((.((..(((.((.(((....))).)...(((((.(((....))).))))).).))).......(((((((...))))))))).))..)))))) ( -44.10) >consensus GG______GAAGCUGGGAG_CUGGGAGCUAAG____GGAGCAGGGGACUGGCGACGUUGGCGGCGAAAGCCUCCGACAUGUGUAAAGGAAUCGUUUAAAAUAAAUGACAGGCGGGC_CCC ......................(((.(((...........(((....)))((...(((((.(((....))).))))).............(((((((...)))))))...)).))).))) (-27.83 = -28.70 + 0.87)

| Location | 19,748,101 – 19,748,199 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -26.80 |
| Energy contribution | -27.05 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
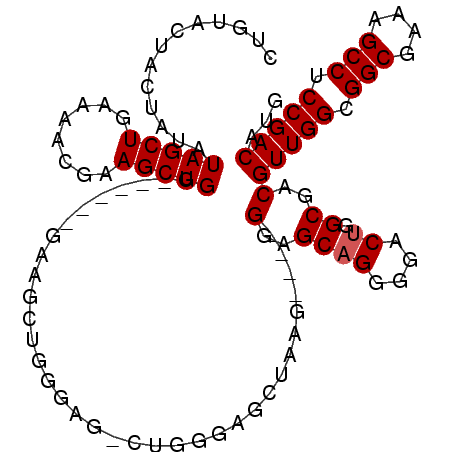
>2R_DroMel_CAF1 19748101 98 - 20766785 CUGUACUACUAUAUAGCUGAAACUGAAGCUG-------GAAGCUGGGAG-CUGGGAGCUAAG----GGAGCAGGGGACUGGCGACGUUGGCGGCGAAAGCCUCCGACAUG .(((.(((((...(((((........)))))-------..)).)))...-...((((((((.----(..((((....)).))..).)))))(((....)))))).))).. ( -34.30) >DroSec_CAF1 890 105 - 1 CUGUACUACUAUAUAGCUGAAAACGAAGCUGGGAGCUGGAAGCUGGGAG-CUGGGAGCUAAG----GGAGCAGGGGACUGGCGACGUUGGCGGCGAAAGCCUCCGACAUG .(((.(((((...(((((........)))))..)).))).(((.....)-)).((((((((.----(..((((....)).))..).)))))(((....)))))).))).. ( -36.90) >DroSim_CAF1 3395 91 - 1 CUGUACUACUAUAUAGCUGAAAACGAAGCUGG--------------GAG-CUGGGAGCUAAG----GGAGCAGGGGACCGGCGACGUUGGCGGCGAAAGCCUCCGACAUG .............(((((........)))))(--------------..(-((((..(((...----..)))......)))))..)(((((.(((....))).)))))... ( -32.40) >DroYak_CAF1 3499 104 - 1 CAGUA---CUAUAUAGCUGAAACUGAAGCUGU-GGCAAGUGGCAGG-GGGUAG-GGGCAUGGGGCAGGGGCAGGGGACUGGCGACGUUGGCGGCGAAAGCCUCCGACAUG ..(..---((((.(..(((...(((..(((..-....)))..))).-...)))-..).))))..)...(.(((....))).)...(((((.(((....))).)))))... ( -36.90) >consensus CUGUACUACUAUAUAGCUGAAAACGAAGCUGG______GAAGCUGGGAG_CUGGGAGCUAAG____GGAGCAGGGGACUGGCGACGUUGGCGGCGAAAGCCUCCGACAUG .............(((((........)))))...................................(..((((....)).))..)(((((.(((....))).)))))... (-26.80 = -27.05 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:43 2006