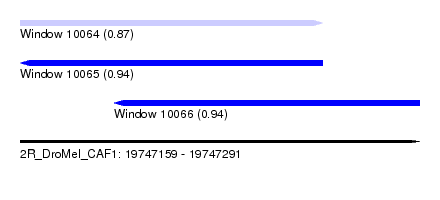
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,747,159 – 19,747,291 |
| Length | 132 |
| Max. P | 0.939055 |

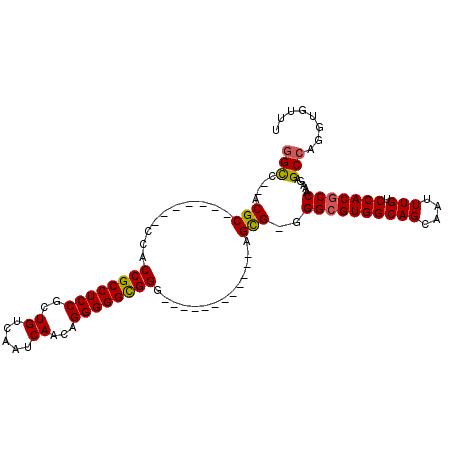
| Location | 19,747,159 – 19,747,259 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.66 |
| Mean single sequence MFE | -43.67 |
| Consensus MFE | -33.49 |
| Energy contribution | -33.42 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19747159 100 + 20766785 GGUC--ACGC-------CCACCGCCUCCGCUGUCAAUCAACAGGGGGUGGGAGCAACAACGGGAGUG-GGGCGUGGCAGCAAUUUGUCCACGCCAAAGAGCCAGGUGUUU ((((--((.(-------((.(((((.((.((((......)))))))))))..(......)))).)))-.((((((((((....))).))))))).....)))........ ( -41.30) >DroSec_CAF1 4 88 + 1 GGCU--GCGC-------CCACCGCCUCCGCUGUCAAUCAACAGGGGGCGGG------------AGCG-GGGAGUGGCAGCAAUUUGUCCACGCCAAAGAGCCAGGUGUUU .(((--((((-------((.((((..(((((.((........)).))))).------------.)))-))).)).))))).........(((((.........))))).. ( -39.10) >DroSim_CAF1 2476 88 + 1 GGCC--ACGC-------CCACCGCCUCCGCUGUCAAUCAACAGGGGGCGGG------------AGCG-GGGCGUGGCAGCAAUUUGUCCACGCCAAAGAGCCAGGUGUUU .(((--((((-------((.(((((.((.((((......))))))))))).------------....-)))))))))............(((((.........))))).. ( -43.60) >DroEre_CAF1 5 104 + 1 CGCCCACCGCCCGCAGCCCACCGCCUCCGCUGUCAAUCAAGAGGGGGCGGGAGG------GGGAGCGAGGGCGUGGCAGCAAUUUGUCCACGCCAAAGAGCCAGGUGUUU ....((((...(((..(((.((((((((.((........)).))))))))...)------))..)))..((((((((((....))).))))))).........))))... ( -50.70) >consensus GGCC__ACGC_______CCACCGCCUCCGCUGUCAAUCAACAGGGGGCGGG____________AGCG_GGGCGUGGCAGCAAUUUGUCCACGCCAAAGAGCCAGGUGUUU (((....(((..........((((((((..((.....))...))))))))..............)))..((((((((((....))).))))))).....)))........ (-33.49 = -33.42 + -0.06)

| Location | 19,747,159 – 19,747,259 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.66 |
| Mean single sequence MFE | -43.75 |
| Consensus MFE | -30.99 |
| Energy contribution | -31.30 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19747159 100 - 20766785 AAACACCUGGCUCUUUGGCGUGGACAAAUUGCUGCCACGCCC-CACUCCCGUUGUUGCUCCCACCCCCUGUUGAUUGACAGCGGAGGCGGUGG-------GCGU--GACC ........((......(((((((.((......))))))))).-.....))...((..(.((((((((((((((.....)))))).)).)))))-------).).--.)). ( -43.40) >DroSec_CAF1 4 88 - 1 AAACACCUGGCUCUUUGGCGUGGACAAAUUGCUGCCACUCCC-CGCU------------CCCGCCCCCUGUUGAUUGACAGCGGAGGCGGUGG-------GCGC--AGCC ........((((....((.((((.((......)))))).)).-((((------------(((((((((((((....))))).)).))))).))-------))).--)))) ( -39.70) >DroSim_CAF1 2476 88 - 1 AAACACCUGGCUCUUUGGCGUGGACAAAUUGCUGCCACGCCC-CGCU------------CCCGCCCCCUGUUGAUUGACAGCGGAGGCGGUGG-------GCGU--GGCC ........((((....(((((((.((......))))))))).-((((------------(((((((((((((....))))).)).))))).))-------))).--)))) ( -45.60) >DroEre_CAF1 5 104 - 1 AAACACCUGGCUCUUUGGCGUGGACAAAUUGCUGCCACGCCCUCGCUCCC------CCUCCCGCCCCCUCUUGAUUGACAGCGGAGGCGGUGGGCUGCGGGCGGUGGGCG ...((((.........(((((((.((......)))))))))(((((..((------(...(((((.((.((........)).)).))))).)))..))))).)))).... ( -46.30) >consensus AAACACCUGGCUCUUUGGCGUGGACAAAUUGCUGCCACGCCC_CGCU____________CCCGCCCCCUGUUGAUUGACAGCGGAGGCGGUGG_______GCGU__GGCC .........((.....(((((((.((......)))))))))...................(((((((((((((.....)))))).)).))))).......))........ (-30.99 = -31.30 + 0.31)

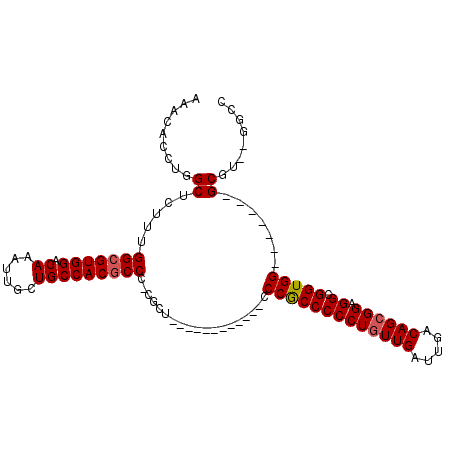
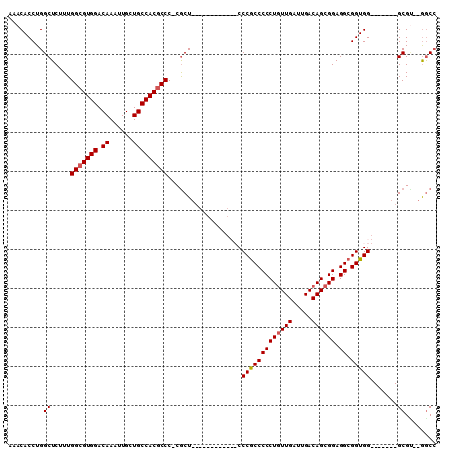
| Location | 19,747,190 – 19,747,291 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.51 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -15.29 |
| Energy contribution | -14.93 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.939055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19747190 101 - 20766785 CAAACUUUGCCGCGGGC-----CAAGGAGA---CAGGCUCA--AACACCUGGCUCUUUGGCGUGGACAAAUU--GCU-GCCAC-----GCCC-CACUCCCGUUGUUGCUCCCACCCCCUG ........((.(((((.-----.((((((.---((((....--....)))).))))))(((((((.((....--..)-)))))-----))).-....))))).))............... ( -34.10) >DroPse_CAF1 8183 108 - 1 AAAACUUUGCCGGGGGCAGCAGCAAAGGCA---GAGGCCAAACAACAUCUGGGUCUUGGACCAGGACAAACACUCCUCUCCACUGCGUGCCC-CACC--UGUC------CGUGGCUCCAG ........((((.((((((..(((..((.(---(((((((.........)))((((((...)))))).......)))))))..)))(((...-))))--))))------).))))..... ( -37.20) >DroSec_CAF1 35 89 - 1 CAAACUUUGCCGCGGGC-----CAAGGAGA---CAGGCUCA--AACACCUGGCUCUUUGGCGUGGACAAAUU--GCU-GCCAC-----UCCC-CGCU------------CCCGCCCCCUG ...........((((((-----(((((((.---((((....--....)))).)))))))))((((.((....--..)-)))))-----...)-))).------------........... ( -34.70) >DroSim_CAF1 2507 89 - 1 CAAACUUUGCCGCGGGC-----CAAGGAGA---CAGGCUCA--AACACCUGGCUCUUUGGCGUGGACAAAUU--GCU-GCCAC-----GCCC-CGCU------------CCCGCCCCCUG ...........(((((.-----(((((((.---((((....--....)))).)))))))((((((.((....--..)-)))))-----))))-))).------------........... ( -35.40) >DroEre_CAF1 45 99 - 1 CAAACUUUGCCGCGGGC-----CAAGGAGCCGCCGGGCUCA--AACACCUGGCUCUUUGGCGUGGACAAAUU--GCU-GCCAC-----GCCCUCGCUCCC------CCUCCCGCCCCCUC ...........(((((.-----...(((((.((((((....--....)))))).....(((((((.((....--..)-)))))-----)))...))))).------...)))))...... ( -43.70) >DroYak_CAF1 2520 77 - 1 CAAACUUUGCCGCGGGC-----CACGAAGA---CGGGCUCA--AACACCUGGCUCUUUGGCGUGGACAAAUU--GCU-GCCAC-----GCC-------------------------CCUG ........((((.((((-----(.((....---))))))).--......)))).....(((((((.((....--..)-)))))-----)))-------------------------.... ( -25.30) >consensus CAAACUUUGCCGCGGGC_____CAAGGAGA___CAGGCUCA__AACACCUGGCUCUUUGGCGUGGACAAAUU__GCU_GCCAC_____GCCC_CGCU____________CCCGCCCCCUG .....(((((((((........(((((((....((((..........)))).))))))).))))).)))).................................................. (-15.29 = -14.93 + -0.36)

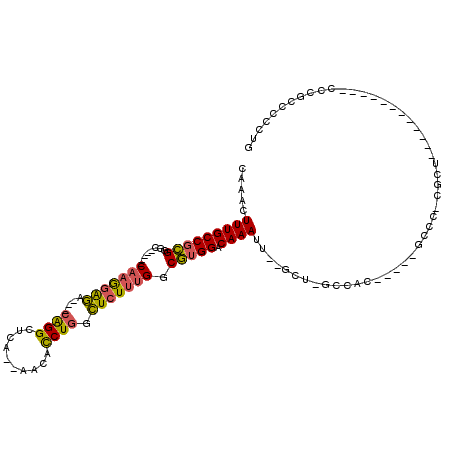
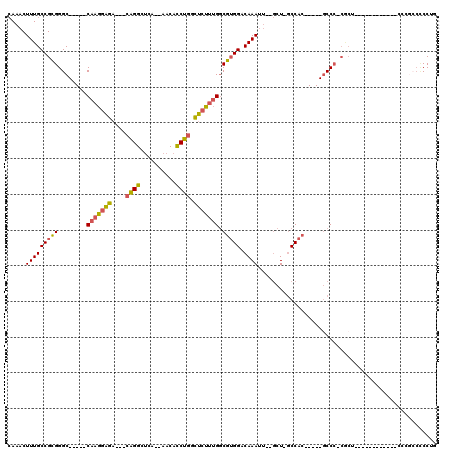
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:39 2006