| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,745,296 – 19,745,457 |
| Length | 161 |
| Max. P | 0.974373 |

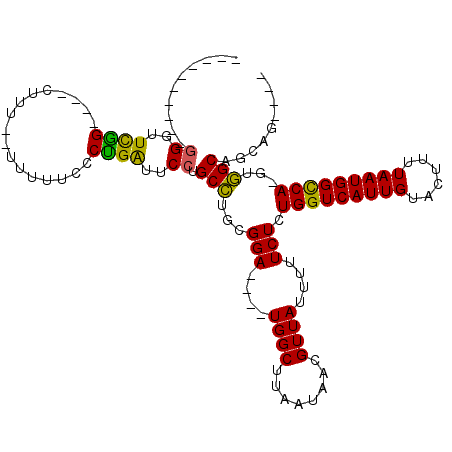
| Location | 19,745,296 – 19,745,387 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.36 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -15.63 |
| Energy contribution | -14.23 |
| Covariance contribution | -1.40 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19745296 91 + 20766785 ----------GGGUUCGG----------UUUUUCCCCGAUUCCUGCCUGCGGA----UGGCUUAAUAACGUUAUUUUUCUCUGGUCAUUGUACUUUUAAUGGCCA-GUGGCAGCAG---- ----------((..((((----------.......))))..))(((.((((((----((((........)))))))..(.((((((((((......)))))))))-).))))))).---- ( -31.00) >DroPse_CAF1 6442 101 + 1 UGUUGUUAUAGGGUUUAG----CUUUCAUUGGUUGCUGGUGCCUGCUU--GGA----UGGCUUAAUAACGUUAUUUUUCUCUGGUCAUUGUACUUUUAAUGGUCAAGCAGC--------- .((((((((((.(((.((----((.((...(((.((....))..))).--.))----.))))....))).)))).......((..(((((......)))))..))))))))--------- ( -24.60) >DroSim_CAF1 615 91 + 1 ----------UGGUUCGG----------UUUUUCCCUGAUUCCUGCCUGCGGA----UGGCUUAAUAACGUUAUUUUUCUCUGGUCAUUGUACUUUUAAUGGCCA-GUGGCAGCAG---- ----------.(..((((----------.......))))..)((((.((((((----((((........)))))))..(.((((((((((......)))))))))-).))))))))---- ( -27.10) >DroYak_CAF1 531 109 + 1 ----------GGGUUCGGUUCGCUUUGGUUUUUCCCUGAUUCCUGCCUGCGGAUGGAUGGCUUAAUAACGUUAUUUUUCUCUGGUCAUUGUACUUUUAAUGGCCA-GUGGCAGCAGGAGC ----------(((.((((......)))).....)))...(((((((.((((((..((((((........))))))..)))((((((((((......)))))))))-)..)))))))))). ( -37.60) >DroPer_CAF1 6444 107 + 1 UGUUGUUAUAGGGUUUAG----CUUUCAUUGGUUGCUGGUGCCUGCUU--GGA----UGGCUUAAUAACGUUAUUUUUCUCUGGUCAUUGUACUUUUAAUGGUCAAGCAGC---AGCAGC (((((((...((((((((----(..(.....)..))))).))))((((--(((----((((........)))))))......(..(((((......)))))..))))))))---)))).. ( -27.80) >consensus __________GGGUUCGG____CUUU__UUUUUCCCUGAUUCCUGCCUGCGGA____UGGCUUAAUAACGUUAUUUUUCUCUGGUCAUUGUACUUUUAAUGGCCA_GUGGCAGCAG____ ..........((..((((.................))))..)).(((...(((....((((........))))....))).(((((((((......)))))))))...)))......... (-15.63 = -14.23 + -1.40)


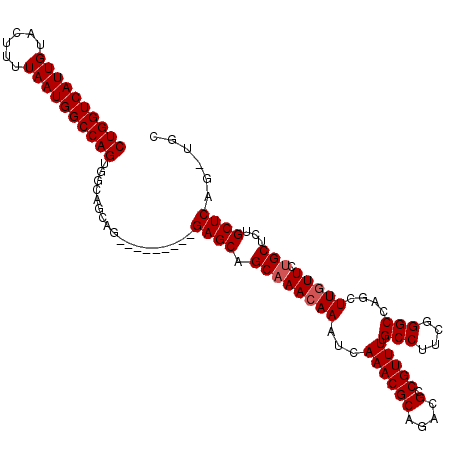
| Location | 19,745,352 – 19,745,457 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 87.69 |
| Mean single sequence MFE | -40.81 |
| Consensus MFE | -26.87 |
| Energy contribution | -29.53 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19745352 105 + 20766785 CUGGUCAUUGUACUUUUAAUGGCCAGUGGCAGCAG---------GAGCAGCAAACAAAUCAAACGCAGACGCCGUUUUGCCUUCGGGCCAGCUUCUUCUGCUCUGCUCAGAUGC ((((((((((......))))))))))(((((((((---------(((.(((.........((((((....).))))).(((....)))..))).)))))))..))).))..... ( -39.90) >DroSim_CAF1 671 105 + 1 CUGGUCAUUGUACUUUUAAUGGCCAGUGGCAGCAG---------GAGCAGCAAACAAAUCAAACGCAGACGCCGUUUUGCCUUCGGGCCAGCUUGUUCUGCGCUGCUCAGCUGC ((((((((((......))))))))))..(((((..---------(((((((.............(((((.((.(((..(((....))).)))..)))))))))))))).))))) ( -45.71) >DroYak_CAF1 601 109 + 1 CUGGUCAUUGUACUUUUAAUGGCCAGUGGCAGCAGGAGCAGCAGGAGCAGCAAACAAAUCAAACGCAGACGCCGUUUUGCCUUAGGGCCAGCUUGUUCAGCUCGGCUC-----U ((((((((((......)))))))))).(((((.(.(.((.((....)).((.............))....))).).)))))..((((((((((.....)))).)))))-----) ( -36.82) >consensus CUGGUCAUUGUACUUUUAAUGGCCAGUGGCAGCAG_________GAGCAGCAAACAAAUCAAACGCAGACGCCGUUUUGCCUUCGGGCCAGCUUGUUCUGCUCUGCUCAG_UGC ((((((((((......))))))))))..................((((.((((((((...((((((....).))))).(((....)))....))))).)))...))))...... (-26.87 = -29.53 + 2.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:32 2006