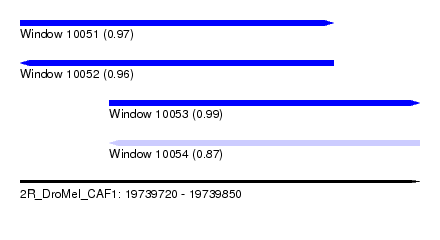
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,739,720 – 19,739,850 |
| Length | 130 |
| Max. P | 0.993480 |

| Location | 19,739,720 – 19,739,822 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.31 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -28.61 |
| Energy contribution | -30.05 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19739720 102 + 20766785 AAACUUU-UUUUGGCCAAAUGGAAUGGAAUGGGUAGCUAUAGCU------GUAGCUAUAGCUUUAGCUGUAGCCCCUGGCUACUGGCUCUCCUGCCCCCCAAAAAGUGG ...(.((-(((((((((.......)))...(((((((((((((.------...))))))))...((((((((((...)))))).))))....))))).)))))))).). ( -38.00) >DroSec_CAF1 49378 101 + 1 AAACUUU-UUUUGGCCAAAUGGAAUGGAAUGGGUACCUAUAGCU------GCAGCUAUAGCUUUGGCUGUAGCCCCUGGCUACUGGCUCUCCUGCCCC-CGAAAAGUGG ..(((((-((.((((((...((......((((....)))).(((------(((((((......)))))))))))).))))))..(((......)))..-.))))))).. ( -35.40) >DroEre_CAF1 49605 102 + 1 AAACUUUUUUUUGGCCAAAUGGAAUGGAAUGGGUAGCUAAAGCU------AUAGCUGCAGCUAUAGCUGUAGCCCCUGGCUACUGGCUCUCCUGCC-CCGAAAAAGUGG ...(.(((((((((((....))...((...((..(((((.((((------(((((....)))))))))((((((...)))))))))))..))..))-))))))))).). ( -40.20) >DroYak_CAF1 50081 108 + 1 AAACUUU-UUUUGGCCAAAUGGAAUGGAAUGGGUAGCUAAAGCUUCAGCAAUAGCUAUAGCUGUAGCUGUAGCCCCUGGCUGCUGGUACUCCUGCCCCCGAAAAUGUGG ..((..(-((((((......((...(((((.((((((((..(((.((((.(((((....))))).)))).)))...)))))))).))..)))..)).))))))).)).. ( -37.30) >consensus AAACUUU_UUUUGGCCAAAUGGAAUGGAAUGGGUAGCUAAAGCU______AUAGCUAUAGCUUUAGCUGUAGCCCCUGGCUACUGGCUCUCCUGCCCCCCAAAAAGUGG ..(((((.....(((.....(((.........((((((((((((........))))))))))))((((((((((...)))))).)))).))).)))......))))).. (-28.61 = -30.05 + 1.44)

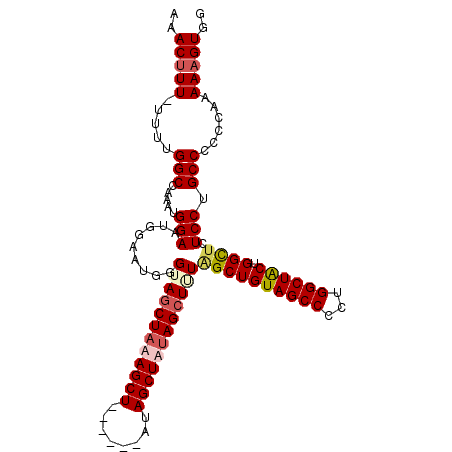
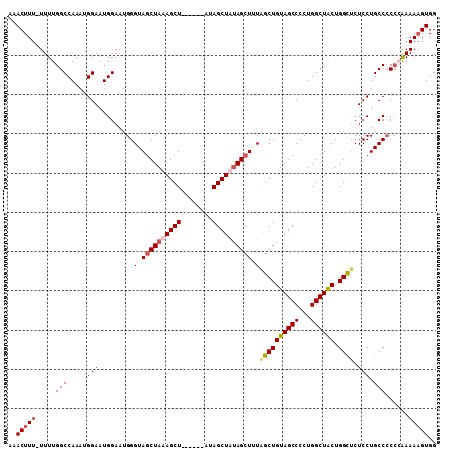
| Location | 19,739,720 – 19,739,822 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.31 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -26.66 |
| Energy contribution | -27.72 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19739720 102 - 20766785 CCACUUUUUGGGGGGCAGGAGAGCCAGUAGCCAGGGGCUACAGCUAAAGCUAUAGCUAC------AGCUAUAGCUACCCAUUCCAUUCCAUUUGGCCAAAA-AAAGUUU ....((((((((((((.((....)).((((((...)))))).))...(((((((((...------.))))))))).)))...(((.......)))))))))-)...... ( -37.60) >DroSec_CAF1 49378 101 - 1 CCACUUUUCG-GGGGCAGGAGAGCCAGUAGCCAGGGGCUACAGCCAAAGCUAUAGCUGC------AGCUAUAGGUACCCAUUCCAUUCCAUUUGGCCAAAA-AAAGUUU ..((((((.(-(((((.((....)).((((((...)))))).)))....(((((((...------.)))))))...)))...(((.......))).....)-))))).. ( -33.10) >DroEre_CAF1 49605 102 - 1 CCACUUUUUCGG-GGCAGGAGAGCCAGUAGCCAGGGGCUACAGCUAUAGCUGCAGCUAU------AGCUUUAGCUACCCAUUCCAUUCCAUUUGGCCAAAAAAAAGUUU ..(((((((...-(((.((..(((..((((((...))))))(((((((((....)))))------))))...)))..))....((.......)))))...))))))).. ( -36.50) >DroYak_CAF1 50081 108 - 1 CCACAUUUUCGGGGGCAGGAGUACCAGCAGCCAGGGGCUACAGCUACAGCUAUAGCUAUUGCUGAAGCUUUAGCUACCCAUUCCAUUCCAUUUGGCCAAAA-AAAGUUU ..........((((((..((((..((((((.....(((((.(((....))).))))).))))))..))))..))).)))...(((.......)))......-....... ( -31.60) >consensus CCACUUUUUCGGGGGCAGGAGAGCCAGUAGCCAGGGGCUACAGCUAAAGCUAUAGCUAC______AGCUAUAGCUACCCAUUCCAUUCCAUUUGGCCAAAA_AAAGUUU ..(((((......(((.((((.....((((((...))))))......((((((((((........))))))))))..........)))).....))).....))))).. (-26.66 = -27.72 + 1.06)

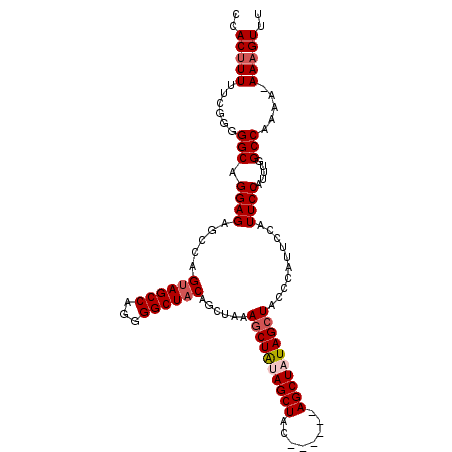
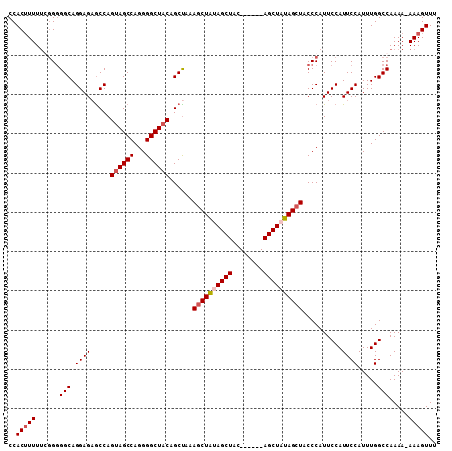
| Location | 19,739,749 – 19,739,850 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 88.62 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -28.66 |
| Energy contribution | -29.10 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19739749 101 + 20766785 GGGUAGCUAUAGCU------GUAGCUAUAGCUUUAGCUGUAGCCCCUGGCUACUGGCUCUCCUGCCCCCCAAAAAGUGGAAACUACGCUAAAAGCUUAAACAAAACC (((((((((((((.------...))))))))...((((((((((...)))))).))))....)))))........(((....).))((.....))............ ( -33.70) >DroSec_CAF1 49407 100 + 1 GGGUACCUAUAGCU------GCAGCUAUAGCUUUGGCUGUAGCCCCUGGCUACUGGCUCUCCUGCCCC-CGAAAAGUGGAAACUACGCUAAAAGCUUAAACAAAACC (((..((....(((------(((((((......))))))))))....)).....(((......)))))-).....(((....).))((.....))............ ( -29.90) >DroEre_CAF1 49635 100 + 1 GGGUAGCUAAAGCU------AUAGCUGCAGCUAUAGCUGUAGCCCCUGGCUACUGGCUCUCCUGCC-CCGAAAAAGUGGAAACUACGCUAAAAGCUUAAACAAAACC (((((((((.((((------(((((....)))))))))((((((...))))))))))).....)))-).......(((....).))((.....))............ ( -33.70) >DroYak_CAF1 50110 107 + 1 GGGUAGCUAAAGCUUCAGCAAUAGCUAUAGCUGUAGCUGUAGCCCCUGGCUGCUGGUACUCCUGCCCCCGAAAAUGUGGAAACUACGCUAAAAGCUUAAACAAAACC (((((((((..(((.((((.(((((....))))).)))).)))...))))))).((((....)))).))......(((....).))((.....))............ ( -32.40) >consensus GGGUAGCUAAAGCU______AUAGCUAUAGCUUUAGCUGUAGCCCCUGGCUACUGGCUCUCCUGCCCCCCAAAAAGUGGAAACUACGCUAAAAGCUUAAACAAAACC .(((((((((((((........)))))))))).((((.((((((...)))))).(((......))).........(((....).))))))..............))) (-28.66 = -29.10 + 0.44)

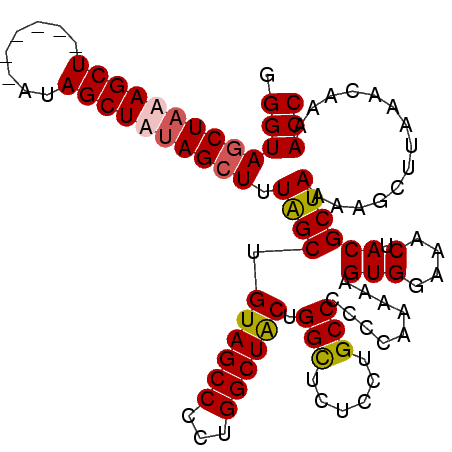
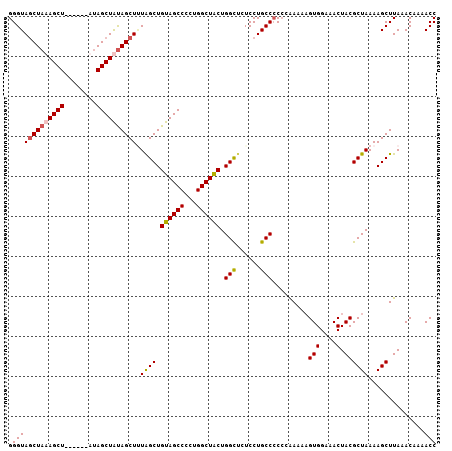
| Location | 19,739,749 – 19,739,850 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.62 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -29.05 |
| Energy contribution | -30.18 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19739749 101 - 20766785 GGUUUUGUUUAAGCUUUUAGCGUAGUUUCCACUUUUUGGGGGGCAGGAGAGCCAGUAGCCAGGGGCUACAGCUAAAGCUAUAGCUAC------AGCUAUAGCUACCC ((((((.((...((.....))...((..((.(.....)))..)))).)))))).((((((...))))))......(((((((((...------.))))))))).... ( -36.70) >DroSec_CAF1 49407 100 - 1 GGUUUUGUUUAAGCUUUUAGCGUAGUUUCCACUUUUCG-GGGGCAGGAGAGCCAGUAGCCAGGGGCUACAGCCAAAGCUAUAGCUGC------AGCUAUAGGUACCC ((((((.((...((.....))...((..((.......)-)..)))).)))))).((((((...))))))........(((((((...------.)))))))...... ( -31.90) >DroEre_CAF1 49635 100 - 1 GGUUUUGUUUAAGCUUUUAGCGUAGUUUCCACUUUUUCGG-GGCAGGAGAGCCAGUAGCCAGGGGCUACAGCUAUAGCUGCAGCUAU------AGCUUUAGCUACCC (((...(((.(((((..((((((((((.((........))-(((.((....)).((((((...)))))).)))..)))))).)))).------))))).))).))). ( -34.10) >DroYak_CAF1 50110 107 - 1 GGUUUUGUUUAAGCUUUUAGCGUAGUUUCCACAUUUUCGGGGGCAGGAGUACCAGCAGCCAGGGGCUACAGCUACAGCUAUAGCUAUUGCUGAAGCUUUAGCUACCC (((........(((((((.((...((..((........))..)).((....))....)).)))))))..(((((.((((.((((....)))).)))).)))))))). ( -35.10) >consensus GGUUUUGUUUAAGCUUUUAGCGUAGUUUCCACUUUUUCGGGGGCAGGAGAGCCAGUAGCCAGGGGCUACAGCUAAAGCUAUAGCUAC______AGCUAUAGCUACCC ((((((.((...((.....))...((..((........))..)))).)))))).((((((...))))))......((((((((((........)))))))))).... (-29.05 = -30.18 + 1.13)

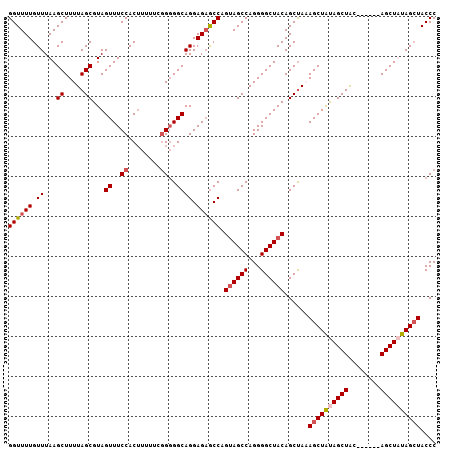
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:27 2006