
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,735,765 – 19,735,856 |
| Length | 91 |
| Max. P | 0.870057 |

| Location | 19,735,765 – 19,735,856 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -16.36 |
| Energy contribution | -15.87 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19735765 91 - 20766785 -------GA------------AUGCUUAUGCCACUUUGCAUAUAAAUCAUUCG--GCAUUGUGGCCAGAACGCAAUGCGCUUAUGAAAUAUUUCGU-UUGUUUAAUGCGCCAA--- -------((------------(((..(((((......))))).....))))).--((((((((.......))))))))((.(((.(((((......-.))))).))).))...--- ( -22.70) >DroVir_CAF1 56841 114 - 1 CUCAUAUUCAGGCUUGAACUCAUAUUCAUAGUACUUUGCAUAUAAAUCAUUUG--GUAUUGUGGCUGAGUCGCAGUGCGCUUAUGAAAUAUUUCGUUUUGUUUAAUGCGCCAGCAA ..........(((..((.((((...((((((((((.((.((....))))...)--))))))))).))))))(((.((.((..(((((....)))))...)).)).))))))..... ( -24.90) >DroPse_CAF1 50387 94 - 1 -------GU------------CUGCUUAUACCACUUUGCAUAUAAAUCAUUUGGGGCAUUGUGGCGAGAGCGCAAUGUGCUUAUGAAAUAUUUCGUUUUGUUUAAUGCGCCAA--- -------..------------...(((...((((..(((...(((.....)))..)))..)))).))).(((((.((.((..(((((....)))))...)).)).)))))...--- ( -21.70) >DroWil_CAF1 69474 82 - 1 ----------------------------AGAUACUUUGCAUAUAAAUCAUUUG--GCAUUGUGGCCAGGACGCAAUGCGCUUAUGAAAUAUUUCGU-UUGUUUAAUGCGGCAA--- ----------------------------.......((((..........((((--((......)))))).((((.((.((..(((((....)))))-..)).)).))))))))--- ( -20.00) >DroAna_CAF1 46050 100 - 1 CUCAUUUGU------------AUGCUUAUGCCACUUUGCAUAUAAAUCAUUUG--GCAUUGUGGCCGAAGCGCAAUGCGCUUAUGAAAUAUUUCGU-UUGUAUAAUGCGCCAAAA- ...((((((------------((((............))))))))))..((((--((......))))))(((((.((..(..(((((....)))))-..)..)).))))).....- ( -28.70) >DroPer_CAF1 46716 94 - 1 -------GU------------CUGCUUAUACCACUUUGCAUAUAAAUCAUUUGGGGCAUUGUGGCGAGAGCGCAAUGUGCUUAUGAAAUAUUUCGUUUUGUUUAAUGCGCCAA--- -------..------------...(((...((((..(((...(((.....)))..)))..)))).))).(((((.((.((..(((((....)))))...)).)).)))))...--- ( -21.70) >consensus _______GU____________AUGCUUAUACCACUUUGCAUAUAAAUCAUUUG__GCAUUGUGGCCAGAGCGCAAUGCGCUUAUGAAAUAUUUCGU_UUGUUUAAUGCGCCAA___ .....................................((((..............((((((((.......))))))))((..(((((....)))))...))...))))........ (-16.36 = -15.87 + -0.50)
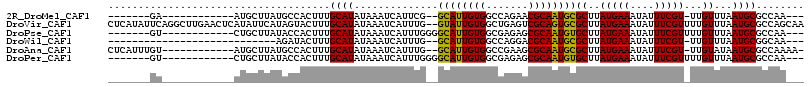
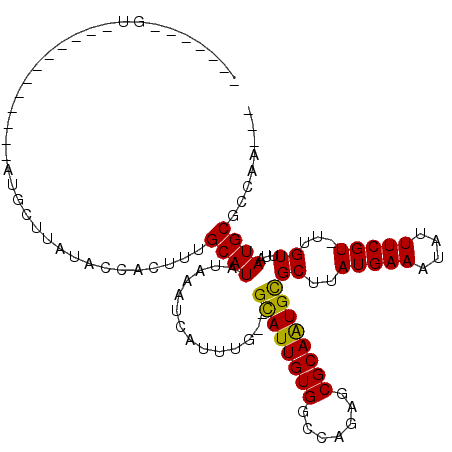

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:22 2006