| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,729,794 – 19,729,908 |
| Length | 114 |
| Max. P | 0.669345 |

| Location | 19,729,794 – 19,729,908 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.67 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -23.67 |
| Energy contribution | -23.12 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
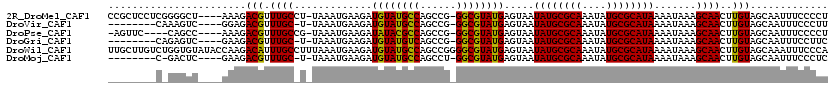
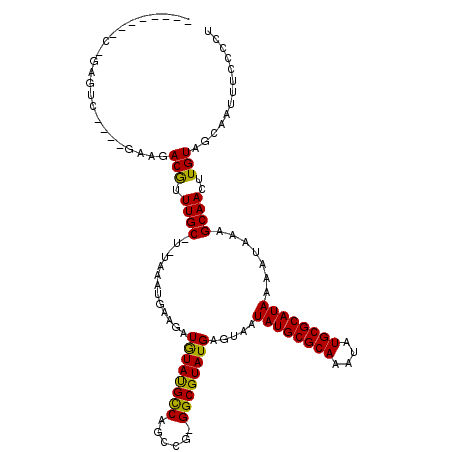
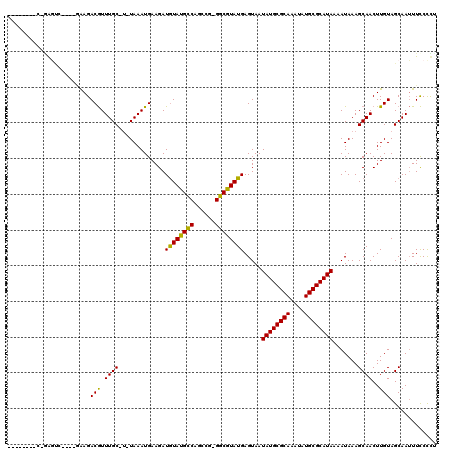
>2R_DroMel_CAF1 19729794 114 + 20766785 CCGCUCCUCGGGGCU----AAAGACGUUUGCCU-UAAAUGAAGAUGUAUGCCAGCCG-GGCGUAUGAGUAAUAUGCGCAAAUAUGCGCAUAAAAUAAAGCAACUUGUAGCAAUUUCCCCU .........((((..----....(((.((((.(-((.........(((((((.....-)))))))......((((((((....))))))))...))).))))..)))........)))). ( -31.74) >DroVir_CAF1 47768 105 + 1 --------CAAAGUC----GGAGACGUUUGC-U-UAAAUGAAGAUGUAUGCCAGCCG-GGCGUAUGAGUAAUAUGCGCAAAUAUGCGCAUAAAAUAAAGCAACUUGUAGCAAUUUCCCUU --------..(((((----(....))..(((-(-(..........(((((((.....-)))))))......((((((((....)))))))).....)))))))))............... ( -28.30) >DroPse_CAF1 44387 109 + 1 -AGUUC----CAGCC----AAAGACGUUUGCCG-UAAAUGAAGAUAUACGCCAGCCG-GGCGUAUGAGUAAUAUGCGCAAAUAUGCGCAUAAAAUAAAGCAACUUGUAGCAAUUUCCCCU -.....----..((.----.(((..((((....-..........((((((((.....-)))))))).....((((((((....)))))))).....))))..)))...)).......... ( -27.90) >DroGri_CAF1 50132 105 + 1 --------CAGAGUC----GAAGACGUUUGC-U-UAAAUGAAGAUGUAUGUCAGCCG-GGCGUAUGAGUAAUAUGCGCAAAUAUGCGCAUAAAAUAAAGCAACUUGUAGCAAUUUCCUUC --------..(((..----....(((.((((-(-(..........(((((((.....-)))))))......((((((((....)))))))).....))))))..)))......))).... ( -24.60) >DroWil_CAF1 60123 120 + 1 UUGCUUGUCUGGUGUAUACCAAGACAUUUGCCUUUAAAUGAAGAUGUAUGCCAGCCGGGGCGUAUGAGUAAUAUGCGCAAAUAUGCGCAUAAAAUAAAGCAACUUGUAGCAAAUUUCCCA (((((((((((((....))).))))).((((.((((.........(((((((......)))))))......((((((((....))))))))...)))))))).....)))))........ ( -35.20) >DroMoj_CAF1 71712 104 + 1 --------C-GACUC----GAAGACGUUUGC-U-UAAAUGAAGAUGUAUGCCAGCCU-GGCGUAUGAGUAAUAUGCGCAAAUAUGCGCAUAAAAUAAAGCAACUUGUAGCAAUUUCCCUC --------.-.....----(...(((.((((-(-(..........(((((((.....-)))))))......((((((((....)))))))).....))))))..)))..).......... ( -27.20) >consensus ________C_GAGUC____GAAGACGUUUGC_U_UAAAUGAAGAUGUAUGCCAGCCG_GGCGUAUGAGUAAUAUGCGCAAAUAUGCGCAUAAAAUAAAGCAACUUGUAGCAAUUUCCCCU .......................(((.((((.............((((((((......)))))))).....((((((((....)))))))).......))))..)))............. (-23.67 = -23.12 + -0.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:21 2006