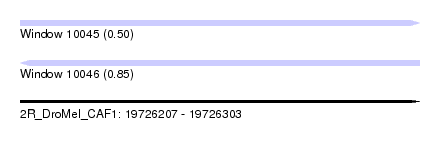
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,726,207 – 19,726,303 |
| Length | 96 |
| Max. P | 0.845754 |

| Location | 19,726,207 – 19,726,303 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -26.99 |
| Consensus MFE | -16.22 |
| Energy contribution | -17.92 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19726207 96 + 20766785 CGGUUCGGUAAAUAAAAUGCCAAAUGGCAAACGGCAAUACCCGAACGCGCAGCUCACCCAUCGAGGGCGAGCUCCAUUGUCGCCAAUUGGCAACUU .................((((((.((((...(((......))).....(.(((((.(((.....))).))))).)......)))).)))))).... ( -33.30) >DroPse_CAF1 40752 72 + 1 --GCUCAGUAAAUAAAAGUGCU--UGGCU---------GCUUG-----------GGUUCUUGGCUGGCUGCCGCCAUUGUCAGCAAUUGGCAACUU --.............(((((((--..(((---------((...-----------(((....(((.....))))))...).))))....))).)))) ( -18.70) >DroSec_CAF1 35933 96 + 1 CGGUUCGGUAAAUAAAAUGCCAAAUGGCAAACGGCAAUACCCGAACGCGCAGCGCAGCCUUCGAGGGCGAGCUCCAUUGUCGCCAAUUGGCAACUU .................((((((.((((...(((......))).....(.(((...((((....))))..))).)......)))).)))))).... ( -29.60) >DroSim_CAF1 41306 96 + 1 CGGUUCGGUAAAUAAAAUGCCAAAUGGCAAACGGCAAUACCCGAACGCGCAGCGCAGCCUUCGAGGGCGAGCUCCAUUGUCGCCAAUUGGCAACUU .................((((((.((((...(((......))).....(.(((...((((....))))..))).)......)))).)))))).... ( -29.60) >DroEre_CAF1 36249 83 + 1 CGGUUCGGUAAAUAAAAUGCCAAAUGGCAAACGGCAAUACCCGAACGCGCAG-------------GGCGAGCUCCAUUGUCGCCAAUUGGCAACUU ..((((((.........((((....))))...........))))))((.((.-------------(((((.(......))))))...))))..... ( -24.15) >DroYak_CAF1 36485 91 + 1 CGGUUCGGUAAAUAAAAUGCCAAAUGGCAAACGGCAAUACCCGAACGCGCAGC-----CCGAGAAGGCGAGCUCCAUUGUCGCCAAUUGGCAACUU .................((((((.((((...(((......)))...((...((-----(......)))..)).........)))).)))))).... ( -26.60) >consensus CGGUUCGGUAAAUAAAAUGCCAAAUGGCAAACGGCAAUACCCGAACGCGCAGC_CA_CCUUCGAGGGCGAGCUCCAUUGUCGCCAAUUGGCAACUU .................((((((.((((...(((......)))......................(((((......))))))))).)))))).... (-16.22 = -17.92 + 1.70)

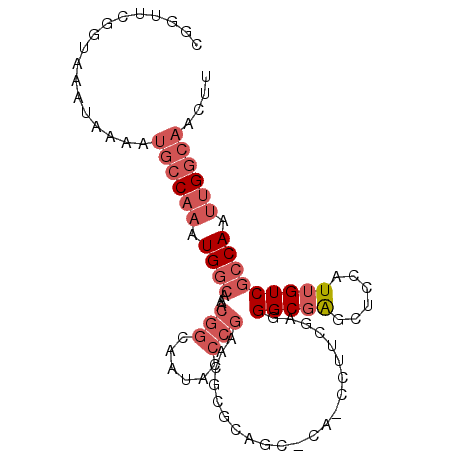
| Location | 19,726,207 – 19,726,303 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -18.47 |
| Energy contribution | -20.05 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19726207 96 - 20766785 AAGUUGCCAAUUGGCGACAAUGGAGCUCGCCCUCGAUGGGUGAGCUGCGCGUUCGGGUAUUGCCGUUUGCCAUUUGGCAUUUUAUUUACCGAACCG ..((((((....))))))...(.(((((((((.....))))))))).)(.((((((...........((((....)))).........))))))). ( -43.85) >DroPse_CAF1 40752 72 - 1 AAGUUGCCAAUUGCUGACAAUGGCGGCAGCCAGCCAAGAACC-----------CAAGC---------AGCCA--AGCACUUUUAUUUACUGAGC-- ..(((((((.(((....)))))))))).(((((.........-----------...((---------.....--.))...........))).))-- ( -13.80) >DroSec_CAF1 35933 96 - 1 AAGUUGCCAAUUGGCGACAAUGGAGCUCGCCCUCGAAGGCUGCGCUGCGCGUUCGGGUAUUGCCGUUUGCCAUUUGGCAUUUUAUUUACCGAACCG ..((((((....))))))...(.(((.((((......))).).))).)(.((((((...........((((....)))).........))))))). ( -33.35) >DroSim_CAF1 41306 96 - 1 AAGUUGCCAAUUGGCGACAAUGGAGCUCGCCCUCGAAGGCUGCGCUGCGCGUUCGGGUAUUGCCGUUUGCCAUUUGGCAUUUUAUUUACCGAACCG ..((((((....))))))...(.(((.((((......))).).))).)(.((((((...........((((....)))).........))))))). ( -33.35) >DroEre_CAF1 36249 83 - 1 AAGUUGCCAAUUGGCGACAAUGGAGCUCGCC-------------CUGCGCGUUCGGGUAUUGCCGUUUGCCAUUUGGCAUUUUAUUUACCGAACCG ....((((((.(((((((....((((.(((.-------------..))).)))).((.....))).)))))).))))))................. ( -27.10) >DroYak_CAF1 36485 91 - 1 AAGUUGCCAAUUGGCGACAAUGGAGCUCGCCUUCUCGG-----GCUGCGCGUUCGGGUAUUGCCGUUUGCCAUUUGGCAUUUUAUUUACCGAACCG ..((((((....))))))...(.((((((......)))-----))).)(.((((((...........((((....)))).........))))))). ( -33.55) >consensus AAGUUGCCAAUUGGCGACAAUGGAGCUCGCCCUCGAAGG_UG_GCUGCGCGUUCGGGUAUUGCCGUUUGCCAUUUGGCAUUUUAUUUACCGAACCG ..((((((....))))))....((((.(((................))).)))).((((........((((....)))).......))))...... (-18.47 = -20.05 + 1.58)

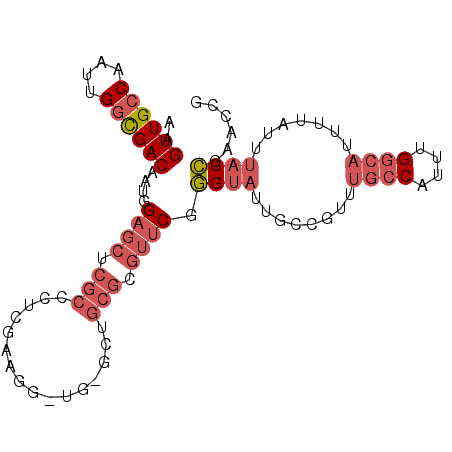
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:19 2006