| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,721,726 – 19,721,830 |
| Length | 104 |
| Max. P | 0.634938 |

| Location | 19,721,726 – 19,721,830 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.79 |
| Mean single sequence MFE | -19.40 |
| Consensus MFE | -12.90 |
| Energy contribution | -12.60 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
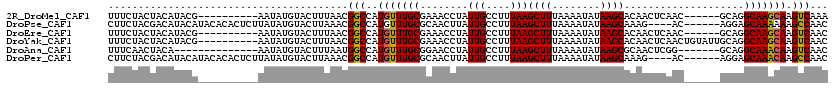
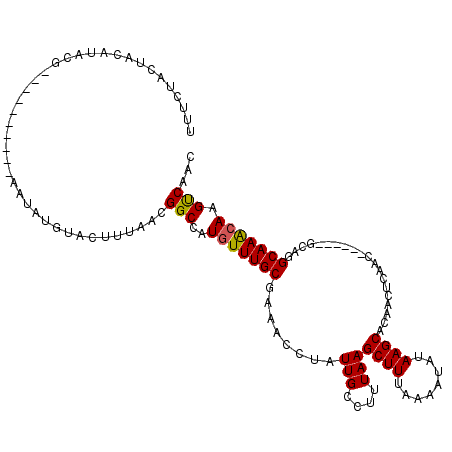
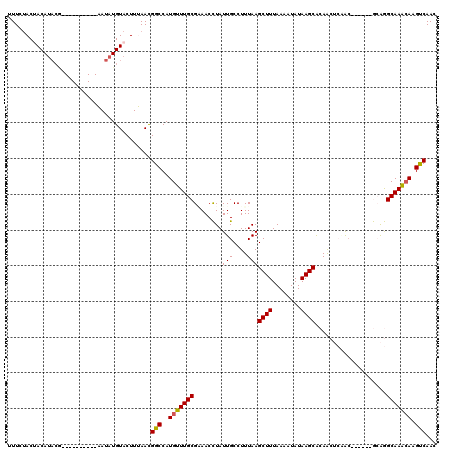
>2R_DroMel_CAF1 19721726 104 - 20766785 UUUCUACUACAUACG----------AAUAUGUACUUUAACGGCCAUGUUUGCGAAACCUAUUGCCUUUAAGCUUUAAAAUAUAAGCACAACUCAAC------GCAGGCAAGCAAGUCAAA .......((((((..----------..)))))).......(((..(((((((..................((((........))))....((....------..))))))))).)))... ( -16.50) >DroPse_CAF1 36092 110 - 1 CUUCUACGACAUACAUACACACUCUUAUAUGUACUUAAACGGCCAUGUUUGCGCAACUUAUUGCCUUUAAGCUUUAAAAUAUAAGCAAAG----AC------AGGAGCAAAAAAGCCAAC ...........((((((..........)))))).......(((....(((((((((....))))((((..((((........))))))))----..------....)))))...)))... ( -18.50) >DroEre_CAF1 31725 104 - 1 UUUCUACUACAUACG----------AAUAUGUACUUUAACGGCCAUGUUUGCGAAACCUAUUGCCUUUAAGCUUUAAAAUAUAAGCACAACUCAAC------GCAGGCAAGCAAGUCAAC .......((((((..----------..)))))).......(((..(((((((..................((((........))))....((....------..))))))))).)))... ( -16.50) >DroYak_CAF1 31837 110 - 1 UUUCUACUACAUACG----------AAUAUGUACUUUAACGGCCAUGUUUGCGAAACCUAUUGCCUUUAAGCUUUAAAAUAUAAGCACAACUCAACUGUAUUGCAGGCAAGCAAGUCAAC .......((((((..----------..)))))).......(((..(((((((..................((((........)))).........(((.....)))))))))).)))... ( -20.50) >DroAna_CAF1 32073 99 - 1 UUUCAACUACA--------------AAUAUGUACUUUAAUGGCCAUGUUUGCGGAACCUAUUGCCUUUAAGCUUUAAAAUAUAAGCGCAACUCGG-------GCAGGCAAACAAGUCAAC ......(..((--------------((((((..(......)..))))))))..)......((((((....((((........))))((.......-------)))))))).......... ( -22.20) >DroPer_CAF1 32342 110 - 1 CUUCUACGACAUACAUACACACUCUUAUAUGUACUUAAACGGCCAUGUUUGCGCAACUUAUUGCCUUUAAGCUUUAAAAUAUAAGCAAAG----AC------AGGAGCAAACAAGCCAAC ...........((((((..........)))))).......(((..(((((((((((....))))((((..((((........))))))))----..------....))))))).)))... ( -22.20) >consensus UUUCUACUACAUACG__________AAUAUGUACUUUAACGGCCAUGUUUGCGAAACCUAUUGCCUUUAAGCUUUAAAAUAUAAGCACAACUCAAC______GCAGGCAAACAAGUCAAC ........................................(((..(((((((........(((....)))((((........))))....................))))))).)))... (-12.90 = -12.60 + -0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:17 2006