| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,697,223 – 19,697,338 |
| Length | 115 |
| Max. P | 0.545219 |

| Location | 19,697,223 – 19,697,338 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -25.36 |
| Energy contribution | -25.12 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
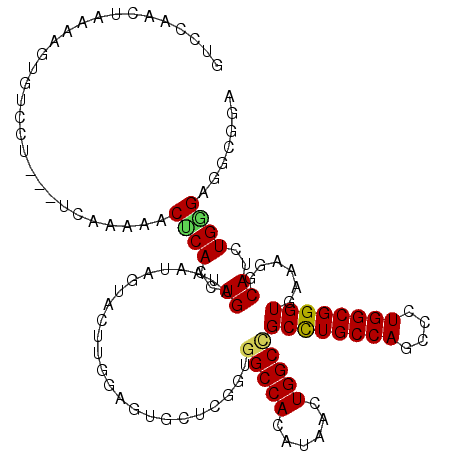
>2R_DroMel_CAF1 19697223 115 - 20766785 GUCCAACGAAAAGUGUCUU---UCAAAAACCCACCUGAUAGUAGUAUUUGGAGUGCUCGGUGGCCACAUAACUGGCUGCCUGCCAGCCCUGGCGGGUAAAAGGCAUUUGGGAGUCGGA .(((.((...(((((((((---(.....((((........(.(((((.....))))))((..((((......))))..)).((((....)))))))).))))))))))....)).))) ( -40.00) >DroPse_CAF1 11309 112 - 1 GGACAGCUGGAA--GGCAU---UCA-AAACUCACCUGAUAAUAAUACUUUGACGUUUCAGUGGCCACAUAGCUGGCCGCCUGCCAGCCCUGGCGGGUGAAAGGCAUCUGAGUGGUGGA .....(((....--))).(---(((-..(((((..((.........((((.((......(((((((......)))))))((((((....)))))))).)))).))..)))))..)))) ( -37.10) >DroEre_CAF1 7185 115 - 1 GUCCAAAUAUAAGUGUCCU---UCAAAAACCCACCUGAUAGUAGUAGUUGGAGUGCUCGGUGGCCACAUAACUGGCCGCCUGCCAACCCUGGCGGGUAAACGGCAUUUGGGAGUCGGA .(((.....((((((((((---((((..((..((......)).))..))))))(((((((((((((......)))))))).((((....)))))))))...))))))))......))) ( -40.00) >DroYak_CAF1 7222 115 - 1 GUCCAAAUAAAAGUGUCCU---UCAAAAACACACCUGAUAGUAGUAGUUGGAGUGCUCGGUGGCCACAUAACUGGCCGCCUGCCAACCUUGGCGGGUGAAUGGCAUUUGGGAGUCGGA .(((......(((((((.(---(((......((((..((.......))..).)))(((((((((((......)))))))).((((....))))))))))).))))))).)))...... ( -39.00) >DroAna_CAF1 7547 118 - 1 UAUCAAGUAAAAGAAACCAGAAGCAAGAACUCACCUGGUAAUAGUACCUGGAAUUCUCCGUGGCCACGUAGCUGGCGGCUUGCCAGCCCUGGCGGGUGAAGGGCAUCUGGGUGGAGGG ................(((((.((.....(((((((((((....)))).(((....)))...((((....((((((.....))))))..))))))))).)).)).)))))........ ( -39.70) >DroPer_CAF1 7696 112 - 1 GGACAGCUGGAA--GGCAU---UCA-AAACUCACCUGAUAAUAAUACUUUGACGUUUCAGUGGCCACAUAGCUGGCCGCCUGCCAGCCCUGGCGGGUGAAAGGCAUCUGAGUGGUGGA .....(((....--))).(---(((-..(((((..((.........((((.((......(((((((......)))))))((((((....)))))))).)))).))..)))))..)))) ( -37.10) >consensus GUCCAACUAAAAGUGUCCU___UCAAAAACUCACCUGAUAAUAGUACUUGGAGUGCUCGGUGGCCACAUAACUGGCCGCCUGCCAGCCCUGGCGGGUGAAAGGCAUCUGGGAGGCGGA .............................((((..((........................(((((......)))))((((((((....))))))))......))..))))....... (-25.36 = -25.12 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:08 2006