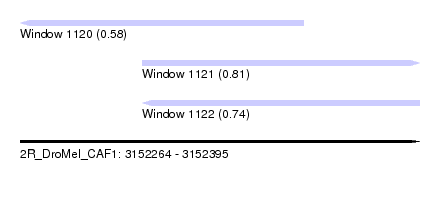
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,152,264 – 3,152,395 |
| Length | 131 |
| Max. P | 0.811719 |

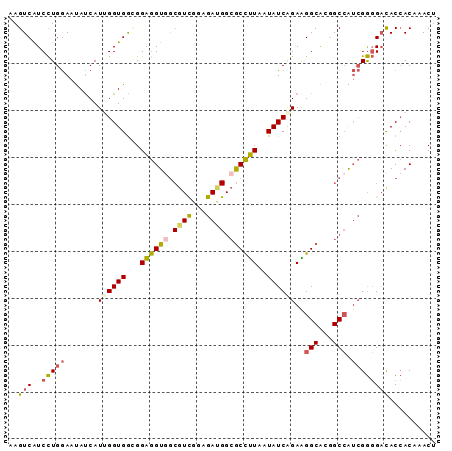
| Location | 3,152,264 – 3,152,357 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 81.86 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -23.04 |
| Energy contribution | -23.43 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3152264 93 - 20766785 CGGAGAAGGCGCCUUUAUAUCAGAAGGCAUGGCCAUCGGGGAUACCACAAACUCAUUGGUUGUCUCUGUGAAUGCGGGCUCAUUCUCCUCCGG (((((.(((.((((((......))))))((((((..(((((((((((.........))).))))))))........))).)))))).))))). ( -32.80) >DroPse_CAF1 38345 84 - 1 ---------AACUAUAAUAUCAGAUGGCGCGGCAGAGGAUGCUACGACGAACGUGUUUGUGGUUUCCGUAAAUGCGGGCUCAUGCUCCUCCGG ---------...............(((.(.(((((((...(((((((((....)).))))))).(((((....)))))))).)))).).))). ( -21.10) >DroSec_CAF1 42004 93 - 1 CGGAGACGGCGCCUUUAUAUCAGAGGGCAUGGCCAUCGGGGACACCACAAACUCAUUGGUUGUCUCCGUGAAUGCGGGCUCGUUCUCCUCCGG .((((((((.((((.(((.(((((.(((...))).))((((((((((.........))).))))))).)))))).))))))).)))))..... ( -36.40) >DroSim_CAF1 44036 93 - 1 CGGAGAUGGCGCCUUUAUAUCAGAAGGCAUGGCCAUCGGGGACACCACAAACUCAUUGGUUGUCUCCGUGAAUGCGGGCUCAUUCUCCUCCGG ((((((((((((((((......))))))...))))))((((((((((.........))).)))))))(.(((((......))))).).)))). ( -38.10) >DroEre_CAF1 41564 93 - 1 UGGAGAUGGCGCCUUAAUAUCAGAAGGCACUGCCAUCGGGGACACCACGAACUCAUUGGUGGUCUCUGUGAAUGCCGGCUCAUUCUCCUCCAG (((((((((((((((........)))))...))))))(((((.(((((.((....)).))))).)))..(((((......))))).)))))). ( -33.60) >DroYak_CAF1 42012 93 - 1 CGGAGAUGGCGCCUUAAUAUCGGAAGGCACUGCCAUCGGGGACACCACGAACUCAUUGGUUGUCUCUGUGAAUGCGGGCUCAUUCUCUUCCAG .((((((((((((((........))))).((((...(((((((((((.........))).)))))))).....)))))).)).)))))..... ( -32.50) >consensus CGGAGAUGGCGCCUUAAUAUCAGAAGGCACGGCCAUCGGGGACACCACAAACUCAUUGGUUGUCUCCGUGAAUGCGGGCUCAUUCUCCUCCGG .(((((.((((((((........))))).(.((...(((((((((((.........)))).))))))).....)).))))...)))))..... (-23.04 = -23.43 + 0.39)

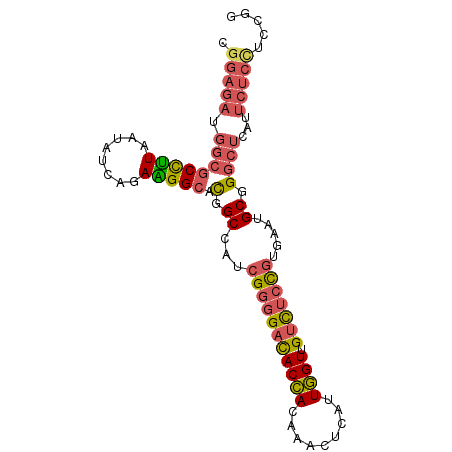
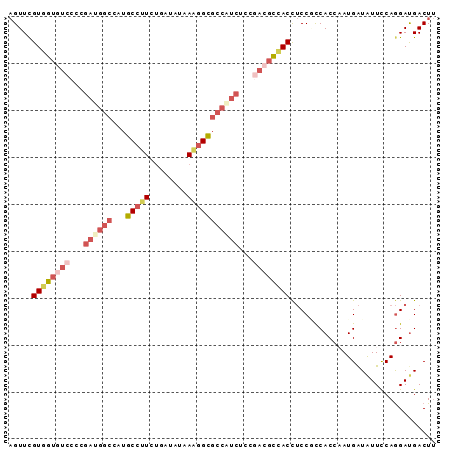
| Location | 3,152,304 – 3,152,395 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -17.96 |
| Energy contribution | -19.93 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3152304 91 + 20766785 AGUUUGUGGUAUCCCCGAUGGCCAUGCCUUCUGAUAUAAAGGCGCCUUCUCCGACGCCACCUCCGCCACCAAUGAUAUUCCAGGAUGACUU ((((.((((..((...((.(((...(((((........)))))))).))...))..)))).(((..................))).)))). ( -20.57) >DroPse_CAF1 38385 79 + 1 CGUUCGUCGUAGCAUCCUCUGCCGCGCCAUCUGAUAUUAUAGUU------------UCACUACCAUCACCCAUGGUGUUUCAAGAUGACUC ...(((((((((......)))).(((((((.((((....(((..------------...)))..))))...))))))).....)))))... ( -16.50) >DroSec_CAF1 42044 91 + 1 AGUUUGUGGUGUCCCCGAUGGCCAUGCCCUCUGAUAUAAAGGCGCCGUCUCCGACGCUACCUCCGCCACCAAUGAUAUUCCAGGAUGACUU ((((.((((((((...((((((...(((............)))))))))...)))))))).(((..................))).)))). ( -23.67) >DroSim_CAF1 44076 91 + 1 AGUUUGUGGUGUCCCCGAUGGCCAUGCCUUCUGAUAUAAAGGCGCCAUCUCCGACGCCACCUCCGCCACCAAUGAUAUUCCAGGAUGACUU ((((.((((((((...((((((...(((((........)))))))))))...)))))))).(((..................))).)))). ( -29.67) >DroEre_CAF1 41604 91 + 1 AGUUCGUGGUGUCCCCGAUGGCAGUGCCUUCUGAUAUUAAGGCGCCAUCUCCAACGCCACCUCCUCCACCCAUGAUAUUCCAGGAUGACUU .....(((((((....((((((...(((((........)))))))))))....)))))))......((.((.((......)))).)).... ( -27.20) >DroYak_CAF1 42052 91 + 1 AGUUCGUGGUGUCCCCGAUGGCAGUGCCUUCCGAUAUUAAGGCGCCAUCUCCGACGCCACCUCCUCCACCAAUGAUAUUCCAGGACGACUU .((((((((((((...((((((...(((((........)))))))))))...))))))))............((......))))))..... ( -31.40) >consensus AGUUCGUGGUGUCCCCGAUGGCCAUGCCUUCUGAUAUAAAGGCGCCAUCUCCGACGCCACCUCCGCCACCAAUGAUAUUCCAGGAUGACUU .....((((((((...((((((...(((((........)))))))))))...))))))))............................... (-17.96 = -19.93 + 1.97)

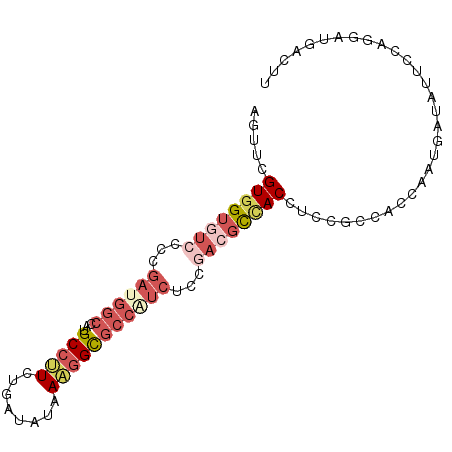
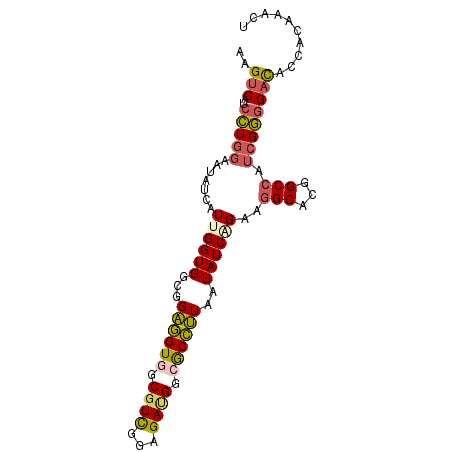
| Location | 3,152,304 – 3,152,395 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.87 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3152304 91 - 20766785 AAGUCAUCCUGGAAUAUCAUUGGUGGCGGAGGUGGCGUCGGAGAAGGCGCCUUUAUAUCAGAAGGCAUGGCCAUCGGGGAUACCACAAACU ..((((((...(.....)...))))))...(((((((((......)))((((((......))))))...))))))((.....))....... ( -30.20) >DroPse_CAF1 38385 79 - 1 GAGUCAUCUUGAAACACCAUGGGUGAUGGUAGUGA------------AACUAUAAUAUCAGAUGGCGCGGCAGAGGAUGCUACGACGAACG ..(((((((.((..((((...))))(((((.....------------.)))))....)))))))))(..(((.....)))..)........ ( -20.50) >DroSec_CAF1 42044 91 - 1 AAGUCAUCCUGGAAUAUCAUUGGUGGCGGAGGUAGCGUCGGAGACGGCGCCUUUAUAUCAGAGGGCAUGGCCAUCGGGGACACCACAAACU ..(((.(((..(.....)...((((((.........((((....))))((((((......))))))...)))))))))))).......... ( -31.20) >DroSim_CAF1 44076 91 - 1 AAGUCAUCCUGGAAUAUCAUUGGUGGCGGAGGUGGCGUCGGAGAUGGCGCCUUUAUAUCAGAAGGCAUGGCCAUCGGGGACACCACAAACU ..((((((...(.....)...))))))....((((.(((...((((((((((((......))))))...))))))...))).))))..... ( -35.10) >DroEre_CAF1 41604 91 - 1 AAGUCAUCCUGGAAUAUCAUGGGUGGAGGAGGUGGCGUUGGAGAUGGCGCCUUAAUAUCAGAAGGCACUGCCAUCGGGGACACCACGAACU ...((((((((......)).)))))).....((((.(((...(((((((((((........)))))...))))))...))).))))..... ( -31.90) >DroYak_CAF1 42052 91 - 1 AAGUCGUCCUGGAAUAUCAUUGGUGGAGGAGGUGGCGUCGGAGAUGGCGCCUUAAUAUCGGAAGGCACUGCCAUCGGGGACACCACGAACU ...((.(((...............))).)).((((.(((...(((((((((((........)))))...))))))...))).))))..... ( -31.86) >consensus AAGUCAUCCUGGAAUAUCAUUGGUGGCGGAGGUGGCGUCGGAGAUGGCGCCUUAAUAUCAGAAGGCACGGCCAUCGGGGACACCACAAACU ..(((..(((((.......((((((...((((((.((((...)))).))))))..))))))..(((...))).)))))))).......... (-19.50 = -19.87 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:08 2006