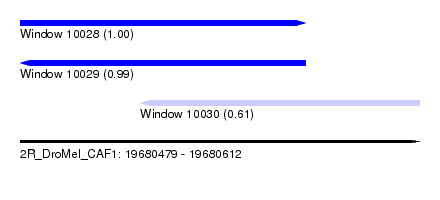
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,680,479 – 19,680,612 |
| Length | 133 |
| Max. P | 0.999580 |

| Location | 19,680,479 – 19,680,574 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.74 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -14.51 |
| Energy contribution | -14.73 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.62 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19680479 95 + 20766785 UAUUCGAAUUCGGUAAGAUCUUAAAGUAUCUUAAAGAUAC------AAAUGCAGUAUGUGUUUACAUACAAAAUCAUACUGCCCUUGAACCCAUU-CAAAGU .....((((..((((((........(((((.....)))))------....(((((((((((......)))....)))))))).)))..))).)))-)..... ( -20.90) >DroSec_CAF1 560 88 + 1 UAU---------CUAAGAU-----AGUAUCUUAAAGAUACUAUCGCAAAUGCAGUAUGUUUUUAGAUACUGAAUUAUACUGCCGUUGAAACCAUUGCAAAGA ..(---------((..(((-----((((((.....)))))))))((((..((((((((..(((((...))))).))))))))...((....))))))..))) ( -25.70) >DroSim_CAF1 1184 88 + 1 CAU---------CUAAGAU-----AGUAUCUUAAAGAUACUAUUGCAUAUGCAGUAUGUUUUUAGAUACUGAAUCAUACUGCCGUUGAAACCAUUGCAAAGA ..(---------((..(((-----((((((.....)))))))))(((...((((((((..(((((...))))).))))))))...((....)).)))..))) ( -23.50) >consensus UAU_________CUAAGAU_____AGUAUCUUAAAGAUACUAU_GCAAAUGCAGUAUGUUUUUAGAUACUGAAUCAUACUGCCGUUGAAACCAUUGCAAAGA ........................((((((.....)))))).........((((((((.(((........))).)))))))).................... (-14.51 = -14.73 + 0.23)

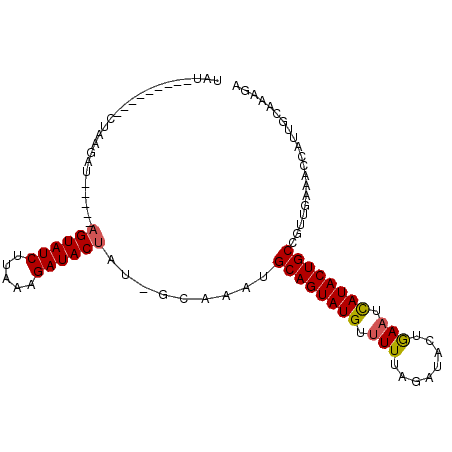
| Location | 19,680,479 – 19,680,574 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 77.74 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -13.12 |
| Energy contribution | -13.46 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19680479 95 - 20766785 ACUUUG-AAUGGGUUCAAGGGCAGUAUGAUUUUGUAUGUAAACACAUACUGCAUUU------GUAUCUUUAAGAUACUUUAAGAUCUUACCGAAUUCGAAUA ..((((-(((.(((..((((((((((((....(((......)))))))))))....------(((((.....))))).......)))))))..))))))).. ( -24.40) >DroSec_CAF1 560 88 - 1 UCUUUGCAAUGGUUUCAACGGCAGUAUAAUUCAGUAUCUAAAAACAUACUGCAUUUGCGAUAGUAUCUUUAAGAUACU-----AUCUUAG---------AUA .....((((..((....)).(((((((..................)))))))..))))(((((((((.....))))))-----)))....---------... ( -20.97) >DroSim_CAF1 1184 88 - 1 UCUUUGCAAUGGUUUCAACGGCAGUAUGAUUCAGUAUCUAAAAACAUACUGCAUAUGCAAUAGUAUCUUUAAGAUACU-----AUCUUAG---------AUG ...(((((...((....)).((((((((................))))))))...)))))(((((((.....))))))-----)......---------... ( -20.39) >consensus UCUUUGCAAUGGUUUCAACGGCAGUAUGAUUCAGUAUCUAAAAACAUACUGCAUUUGC_AUAGUAUCUUUAAGAUACU_____AUCUUAG_________AUA ....................((((((((................))))))))..........(((((.....)))))......................... (-13.12 = -13.46 + 0.33)

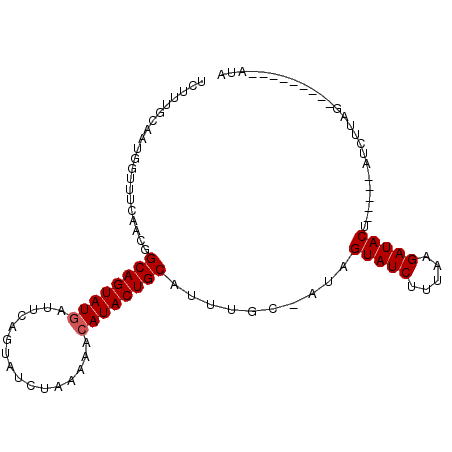
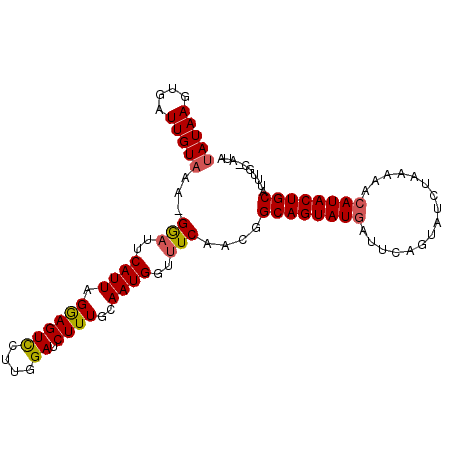
| Location | 19,680,519 – 19,680,612 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.12 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19680519 93 - 20766785 AAUAAGUGAUUGUAAA-GAAUUCAUUAGGAGUCAAUGGAACUUUG-AAUGGGUUCAAGGGCAGUAUGAUUUUGUAUGUAAACACAUACUGCAUUU------ .((((....))))...-(((((((((..(((((....).))))..-)))))))))....((((((((....(((......)))))))))))....------ ( -24.10) >DroSec_CAF1 586 100 - 1 UAUAAGUGAUUGUAAA-GGAUUCAUUAGGAGUUCUUGGAUCUUUGCAAUGGUUUCAACGGCAGUAUAAUUCAGUAUCUAAAAACAUACUGCAUUUGCGAUA ........((((((((-(.(((((...........)))))))))))))).(((.(((..(((((((..................)))))))..))).))). ( -20.07) >DroSim_CAF1 1210 101 - 1 UAUAAGUGAUUGUAAAAGGUCUCAUUAGAAGUCCUUGGAUCUUUGCAAUGGUUUCAACGGCAGUAUGAUUCAGUAUCUAAAAACAUACUGCAUAUGCAAUA ........((((((...((..(((((.((((((....)).))))..)))))..))....((((((((................))))))))...)))))). ( -22.19) >consensus UAUAAGUGAUUGUAAA_GGAUUCAUUAGGAGUCCUUGGAUCUUUGCAAUGGUUUCAACGGCAGUAUGAUUCAGUAUCUAAAAACAUACUGCAUUUGC_AUA (((((....)))))...(((..((((.((((((....)).))))..))))..)))....((((((((................)))))))).......... (-15.78 = -16.12 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:03 2006