
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,662,529 – 19,662,661 |
| Length | 132 |
| Max. P | 0.849981 |

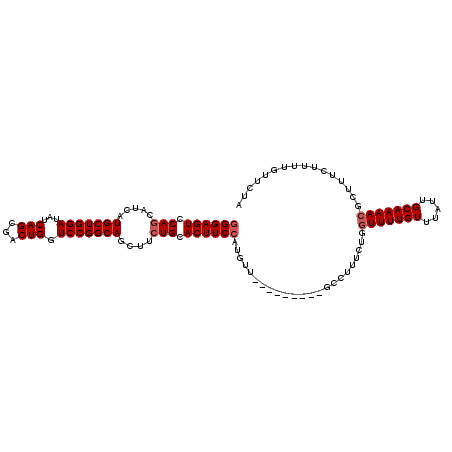
| Location | 19,662,529 – 19,662,634 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 88.59 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -24.33 |
| Energy contribution | -25.33 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
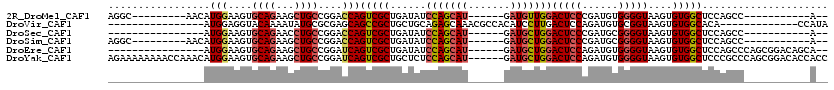
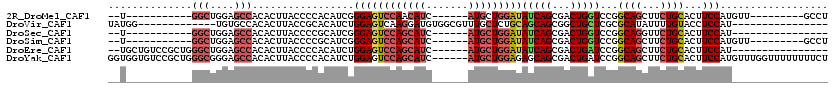
>2R_DroMel_CAF1 19662529 105 - 20766785 GGGAGUCCAACAUCAUGCUGGAUAUCAGCGACUGGUCCGGCAGCUUCUGCACUUCCAUGUU---------GCCUUUCUGGUUUUGUUUAUUGCAAAACGCUUUCUUUUGUUCUA (..(((.((((((..((((((((..(((...)))))))))))((....))......)))))---------)........(((((((.....))))))))))..).......... ( -29.00) >DroSim_CAF1 702 105 - 1 GGGAGUCCAGCAUCAUGCUGGAUAUCAGCGACUGGUCCGGCAGCUUCUGCACUUCCAUGUU---------GCCUUUCUGGUUUUGUUUAUUGCAAAACGCUUUCUUUUGUUCUU (..((((((((.....)))))).....((((((((....((((...))))....))).)))---------))))..)..(((((((.....)))))))................ ( -33.70) >DroYak_CAF1 772 114 - 1 UGGAGUCCAGCAUCAUGCUGGAGAGCAGCGACUGAUCCGGCAGCUUCUGCACUUCCAUGUUUGGUUUUUUUUCUUGCUGCUUUUGUUUAUUGCAAAACGCUUUCUUUUGUUCUA (((((((((((.....))))))(((.((((........((((((..........(((....)))...........))))))(((((.....))))).))))))).....))))) ( -31.70) >consensus GGGAGUCCAGCAUCAUGCUGGAUAUCAGCGACUGGUCCGGCAGCUUCUGCACUUCCAUGUU_________GCCUUUCUGGUUUUGUUUAUUGCAAAACGCUUUCUUUUGUUCUA ((((((.(((.....(((((((...(((...))).)))))))....))).)))))).......................(((((((.....)))))))................ (-24.33 = -25.33 + 1.00)


| Location | 19,662,569 – 19,662,661 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.03 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -20.67 |
| Energy contribution | -21.12 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
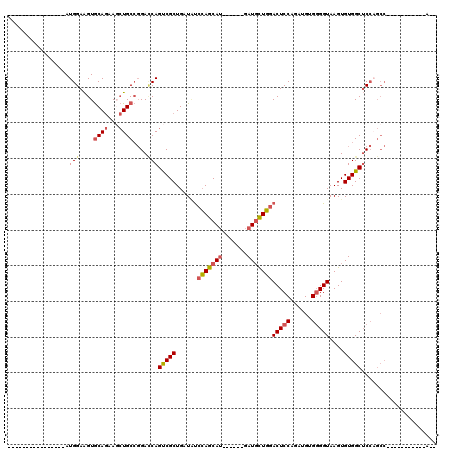
>2R_DroMel_CAF1 19662569 92 + 20766785 AGGC---------AACAUGGAAGUGCAGAAGCUGCCGGACCAGUCGCUGAUAUCCAGCAU------GAUGUUGGACUCCCGAUGUGGGGUAAGUGUGGCUCCAGCC-----------A-- .(((---------.((((((.(((.(((..((((......))))..)))....(((((..------...)))))))).)).))))(((((.......))))).)))-----------.-- ( -31.50) >DroVir_CAF1 1 91 + 1 ----------------AUGGAGGUACAAAAUAUGCGCGAGCAGCCGCUGCUGCAGAGCAAACGCCACAUCCUUGACUCCAGAUGUGCGGUAAGUGUGGCACA-------------CCAUA ----------------((((..((........(((....)))(((((((((....))))..(((.(((((..((....))))))))))......))))))).-------------)))). ( -29.50) >DroSec_CAF1 1 85 + 1 ----------------AUGGAAGUGCAGAACCUGCCGGACCAGUCGCUGAUAUCCAGCAU------GAUGCUGGACUCCCGAUGCGGGGUAAGUGUGGCUCCAGCC-----------A-- ----------------.(((..(.((((...)))))((((((..((((....((((((..------...)))))).(((((...)))))..))))))).)))..))-----------)-- ( -30.10) >DroSim_CAF1 742 92 + 1 AGGC---------AACAUGGAAGUGCAGAAGCUGCCGGACCAGUCGCUGAUAUCCAGCAU------GAUGCUGGACUCCCGAUGCGGGGUAAGUGUGGCUCCAGCC-----------A-- .(((---------.....((.(((......))).))((((((..((((....((((((..------...)))))).(((((...)))))..))))))).))).)))-----------.-- ( -34.70) >DroEre_CAF1 1 96 + 1 ----------------AUGGAAGUGCAGAAGCUGCCGGAUCAGUCGCUGAUAUCCAGCAU------GAUGCUGGACUCCAGAUGUGGGGUAAGUGUGGCUCCAGCCCAGCGGACAGCA-- ----------------..............((((((..((((...((((.....)))).)------)))((((.((((((....))))))......(((....))))))))).)))).-- ( -33.50) >DroYak_CAF1 812 114 + 1 AGAAAAAAAACCAAACAUGGAAGUGCAGAAGCUGCCGGAUCAGUCGCUGCUCUCCAGCAU------GAUGCUGGACUCCAGAUGUGGGGUAAGUGUGGCUCCCGCCCAGCGGACACCACC .........(((..(((((((.(.((((..((((......))))..)))).)((((((..------...)))))).))))..)))..)))....((((...((((...))))...)))). ( -40.30) >consensus ________________AUGGAAGUGCAGAAGCUGCCGGACCAGUCGCUGAUAUCCAGCAU______GAUGCUGGACUCCAGAUGUGGGGUAAGUGUGGCUCCAGCC___________A__ .................(((....((((...))))....)))(((((......(((((((.......)))))))(((((......)))))....)))))..................... (-20.67 = -21.12 + 0.45)
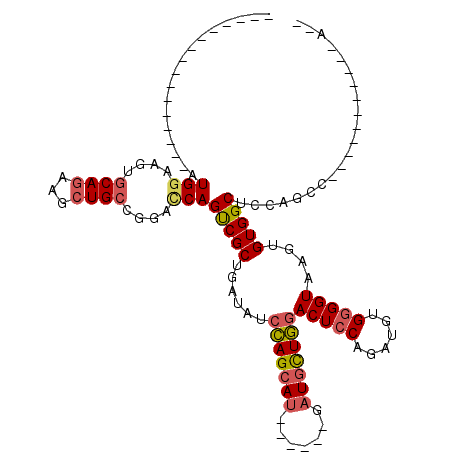
| Location | 19,662,569 – 19,662,661 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.03 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -18.12 |
| Energy contribution | -19.27 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19662569 92 - 20766785 --U-----------GGCUGGAGCCACACUUACCCCACAUCGGGAGUCCAACAUC------AUGCUGGAUAUCAGCGACUGGUCCGGCAGCUUCUGCACUUCCAUGUU---------GCCU --(-----------(((....)))).......(((.....))).(((((.(...------..).)))))....((((((((....((((...))))....))).)))---------)).. ( -28.00) >DroVir_CAF1 1 91 - 1 UAUGG-------------UGUGCCACACUUACCGCACAUCUGGAGUCAAGGAUGUGGCGUUUGCUCUGCAGCAGCGGCUGCUCGCGCAUAUUUUGUACCUCCAU---------------- ...((-------------(((((..........)))))))(((((...(((((((((((...((.((((....))))..)).))).))))))))....))))).---------------- ( -32.80) >DroSec_CAF1 1 85 - 1 --U-----------GGCUGGAGCCACACUUACCCCGCAUCGGGAGUCCAGCAUC------AUGCUGGAUAUCAGCGACUGGUCCGGCAGGUUCUGCACUUCCAU---------------- --(-----------(((....))))...............((((((.(((.(((------.((((((((..(((...)))))))))))))).))).))))))..---------------- ( -31.80) >DroSim_CAF1 742 92 - 1 --U-----------GGCUGGAGCCACACUUACCCCGCAUCGGGAGUCCAGCAUC------AUGCUGGAUAUCAGCGACUGGUCCGGCAGCUUCUGCACUUCCAUGUU---------GCCU --(-----------(((....)))).......((((...)))).(((((((...------..)))))))....((((((((....((((...))))....))).)))---------)).. ( -35.40) >DroEre_CAF1 1 96 - 1 --UGCUGUCCGCUGGGCUGGAGCCACACUUACCCCACAUCUGGAGUCCAGCAUC------AUGCUGGAUAUCAGCGACUGAUCCGGCAGCUUCUGCACUUCCAU---------------- --.((((((((((((((....))).........(((....))).(((((((...------..)))))))..)))))........))))))..............---------------- ( -31.80) >DroYak_CAF1 812 114 - 1 GGUGGUGUCCGCUGGGCGGGAGCCACACUUACCCCACAUCUGGAGUCCAGCAUC------AUGCUGGAGAGCAGCGACUGAUCCGGCAGCUUCUGCACUUCCAUGUUUGGUUUUUUUUCU .(((((..((((...))))..))))).......(((.((.(((((((((((...------..))))))..((((...(((......)))...))))..))))).)).))).......... ( -41.00) >consensus __U___________GGCUGGAGCCACACUUACCCCACAUCGGGAGUCCAGCAUC______AUGCUGGAUAUCAGCGACUGGUCCGGCAGCUUCUGCACUUCCAU________________ ..............(((....))).................((((((((((((.......)))))))))(((((...)))))...((((...))))...))).................. (-18.12 = -19.27 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:56 2006