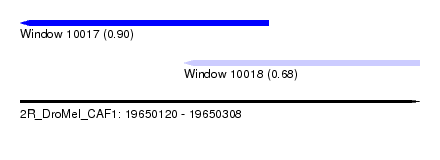
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,650,120 – 19,650,308 |
| Length | 188 |
| Max. P | 0.904300 |

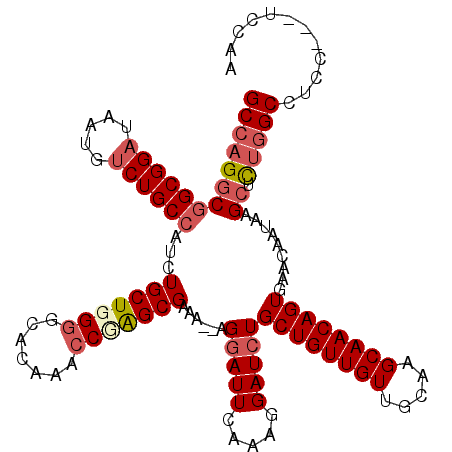
| Location | 19,650,120 – 19,650,237 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.88 |
| Mean single sequence MFE | -41.38 |
| Consensus MFE | -32.50 |
| Energy contribution | -34.25 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19650120 117 - 20766785 GCCAGGCGGCGGAUAAUGUCUGCCAUCUGCUGGGGCGCAAACCUAGCGAAA--AGGAUUCAAAGGAUCUGCUGUUGUUGCAAGCAACAGUGAACAAUAAGCUUUGGCCUCAUCCUCCAA ((((((((((((((...)))))))...(((((((.......)))))))...--.(((((.....)))))((((((((.....)))))))).........)).)))))............ ( -41.70) >DroSec_CAF1 6887 117 - 1 GCCAGGCGGCGGAUAAUGCCUGCCAUCUGCUGGGGCGCAAACCCAGCGAAA--AGGAUUCAAAGGAUAUGCUGUUGUUGCAAGCAACAGUGAACAAUAAGCUCUGGCCUCCUCCUCCAA ((((((((((((.......)))))((((((((((.......))))))(((.--....)))...))))..((((((((.....)))))))).........)).)))))............ ( -43.50) >DroEre_CAF1 6943 115 - 1 GCCAGGCGGCGGAUAAUGUCUGCCAUCUGCUGGGGCACAAACCGGGCGAAAAGAGGAUUCCCAGGAUCUGCUGUUGUUGCAAGCAACAGUGAACAAUGAGCUCCAGCCCC----UUCCA ...(((.(((((((...)))))))....((((((((.((..((.((.(((.......))))).)).((.((((((((.....))))))))))....)).)))))))).))----).... ( -42.60) >DroYak_CAF1 7155 116 - 1 GCCAGCCAGCGGAUAAUGUCUGCCAUCUGCUGGGACAGAAGACGGGCGAAAAAAGGAUUCAAAGGAUCUGCUGUUGUUGCAAGCAACAGUGAACAAUGAGCUCUGGCCUCC---UCCUA ((((.(((((((((..........)))))))))..........((((.......(((((.....)))))((((((((.....)))))))).........))))))))....---..... ( -37.70) >consensus GCCAGGCGGCGGAUAAUGUCUGCCAUCUGCUGGGGCACAAACCGAGCGAAA__AGGAUUCAAAGGAUCUGCUGUUGUUGCAAGCAACAGUGAACAAUAAGCUCUGGCCUCC___UCCAA (((((((((((((.....))))))...(((((((.......)))))))......(((((.....)))))((((((((.....)))))))).........)).)))))............ (-32.50 = -34.25 + 1.75)

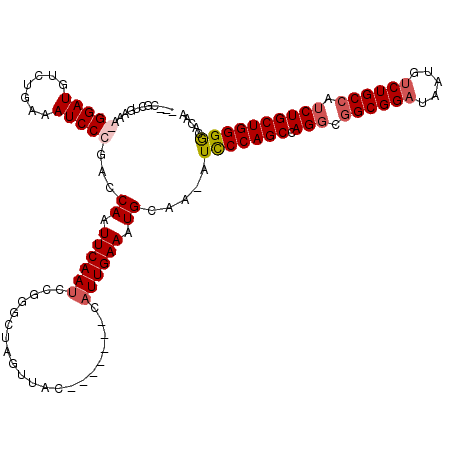
| Location | 19,650,197 – 19,650,308 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.89 |
| Mean single sequence MFE | -35.33 |
| Consensus MFE | -26.33 |
| Energy contribution | -27.45 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19650197 111 - 20766785 CACCGCUGAAA-GGAUGUCUGAAAUCCCGACCAAUUCAAUCCGGGCUAGUUAC-------CAUUGAAAUGCAA-AUCCCAGCCAGGCGGCGGAUAAUGUCUGCCAUCUGCUGGGGCGCAA ...(((.....-((((.......))))....((.((((((..((........)-------))))))).))...-..((((((.((..(((((((...)))))))..)))))))))))... ( -35.70) >DroSec_CAF1 6964 100 - 1 ------------GGAUGUCUGAAAUCCCGACCAAUUCAAUCCGGGCUAGUUAC-------CAUUGAAAUGCAA-AUCCCAGCCAGGCGGCGGAUAAUGCCUGCCAUCUGCUGGGGCGCAA ------------((((.......)))).......((((((..((........)-------))))))).(((..-.(((((((.((..(((((.......)))))..))))))))).))). ( -33.10) >DroEre_CAF1 7018 120 - 1 CGCCGCUGAAAGGGAUGUCUGAAAUCCCGACCAAUUCAAACCGGGCUAGUUACUGAAAUGCAUUGAAAUGCAAAAUCCCAGCCAGGCGGCGGAUAAUGUCUGCCAUCUGCUGGGGCACAA .(((((.....(((((.......)))))...((((.((...(((........)))...)).))))....))......(((((.((..(((((((...)))))))..)))))))))).... ( -40.70) >DroYak_CAF1 7231 110 - 1 ---CGCUGAAAGGGAUGUCUGAAAUCCCGACCAAUUCAAUCCGGGCUAGUUAC-------CAUUGAAAUGCAAAAUUCCAGCCAGCCAGCGGAUAAUGUCUGCCAUCUGCUGGGACAGAA ---..(((...(((((.......)))))...((.((((((..((........)-------))))))).)).....(((((((.((...((((((...))))))...)))))))))))).. ( -31.80) >consensus ___CGCUGAAA_GGAUGUCUGAAAUCCCGACCAAUUCAAUCCGGGCUAGUUAC_______CAUUGAAAUGCAA_AUCCCAGCCAGGCGGCGGAUAAUGUCUGCCAUCUGCUGGGGCACAA ...........(((((.......)))))...((.((((((.....................)))))).)).....(((((((.(((.((((((.....)))))).))))))))))..... (-26.33 = -27.45 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:52 2006