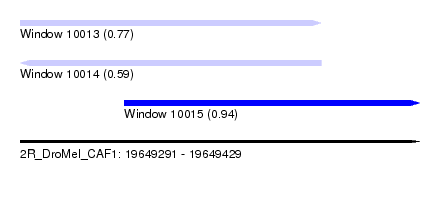
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,649,291 – 19,649,429 |
| Length | 138 |
| Max. P | 0.937573 |

| Location | 19,649,291 – 19,649,395 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.28 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -24.91 |
| Energy contribution | -25.69 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19649291 104 + 20766785 UCCUGACUACGUGCCGCACCAAAGCCCUUCCACGAU------GAUUACGUUGUGAAGGAGCACGCCACUCCGGACUCCGCA-------CUCCCGAGUCGCCCAAGUGUUUAAAACAC ...(((.(((((((.((......))(((((.(((((------(....))))))))))).))))((.((((.(((.......-------.))).)))).))....))).)))...... ( -29.80) >DroEre_CAF1 6142 104 + 1 UCCUGACUACGUGCCGCACAAAAGCCCUUCCACGAU------GAUUACGUUGUGAAGGAGCACGCCACUCCGGACUCCGUG-------CUCCGGAGUCCCCCAAGUGUUUAAAACAC .........(((((.((......))(((((.(((((------(....))))))))))).))))).((((..((((((((..-------...))))))))....)))).......... ( -36.70) >DroYak_CAF1 6289 117 + 1 UCCUGACUACGUGCCACACAAAGCCCCUUCCACGAUGAUGAUGAUUACGUUGUGAAGGAGCACGCCACUCCGGACUCCGUUCCACGGUCUCCGGAGUCGCCCAAGUGUUUAAAACAC ..........((...((((...((.(((((.((((((..........))))))))))).))..((.((((((((..(((.....)))..)))))))).))....)))).....)).. ( -41.40) >consensus UCCUGACUACGUGCCGCACAAAAGCCCUUCCACGAU______GAUUACGUUGUGAAGGAGCACGCCACUCCGGACUCCGUA_______CUCCGGAGUCGCCCAAGUGUUUAAAACAC ..........((...((((....(((((((.(((((............)))))))))).))..((.((((((((...............)))))))).))....)))).....)).. (-24.91 = -25.69 + 0.78)

| Location | 19,649,291 – 19,649,395 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.28 |
| Mean single sequence MFE | -42.27 |
| Consensus MFE | -34.24 |
| Energy contribution | -34.69 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19649291 104 - 20766785 GUGUUUUAAACACUUGGGCGACUCGGGAG-------UGCGGAGUCCGGAGUGGCGUGCUCCUUCACAACGUAAUC------AUCGUGGAAGGGCUUUGGUGCGGCACGUAGUCAGGA ............(((((((.((((.(((.-------(....).))).)))).))((((((((((...(((.....------..))).)))))((......)))))))....))))). ( -37.10) >DroEre_CAF1 6142 104 - 1 GUGUUUUAAACACUUGGGGGACUCCGGAG-------CACGGAGUCCGGAGUGGCGUGCUCCUUCACAACGUAAUC------AUCGUGGAAGGGCUUUUGUGCGGCACGUAGUCAGGA ..........(((((...((((((((...-------..)))))))).)))))((((((((((((...(((.....------..))).)))))((......)))))))))........ ( -42.40) >DroYak_CAF1 6289 117 - 1 GUGUUUUAAACACUUGGGCGACUCCGGAGACCGUGGAACGGAGUCCGGAGUGGCGUGCUCCUUCACAACGUAAUCAUCAUCAUCGUGGAAGGGGCUUUGUGUGGCACGUAGUCAGGA (((((....((((....((.((((((((..(((.....)))..)))))))).))..((((((((...(((.............))).))))))))...))))))))).......... ( -47.32) >consensus GUGUUUUAAACACUUGGGCGACUCCGGAG_______AACGGAGUCCGGAGUGGCGUGCUCCUUCACAACGUAAUC______AUCGUGGAAGGGCUUUUGUGCGGCACGUAGUCAGGA ..........(((((.((..((((((............)))))))).)))))((((((((((((...(((.............))).)))))((......)))))))))........ (-34.24 = -34.69 + 0.45)

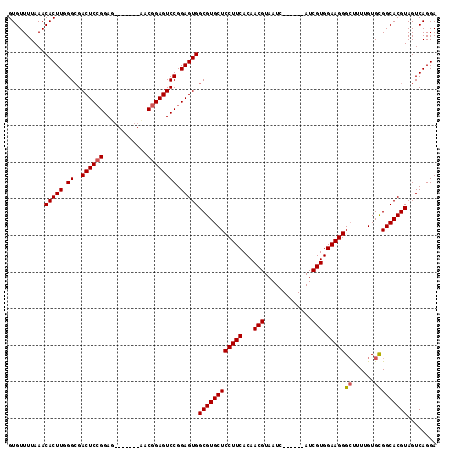
| Location | 19,649,327 – 19,649,429 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -19.73 |
| Energy contribution | -20.73 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19649327 102 + 20766785 --GAUUACGUUGUGAAGGAGCACGCCACUCCGGACUCCGCA-------CUCCCGAGUCGCCCAAGUGUUUAAAACACCCUUCAGCCUUCAGCCCUCAGACUUCAGACC-CCC --......((((((((((.....((.((((.(((.......-------.))).)))).))....((((.....)))))))))).....))))................-... ( -21.60) >DroEre_CAF1 6178 90 + 1 --GAUUACGUUGUGAAGGAGCACGCCACUCCGGACUCCGUG-------CUCCGGAGUCCCCCAAGUGUUUAAAACACCCUGCAGCC-----CU--------UCAGACCUCCC --......(((.((((((.((..((......((((((((..-------...)))))))).....((((.....))))...)).)))-----))--------))))))..... ( -29.30) >DroYak_CAF1 6329 93 + 1 AUGAUUACGUUGUGAAGGAGCACGCCACUCCGGACUCCGUUCCACGGUCUCCGGAGUCGCCCAAGUGUUUAAAACACCCUUCAGCC------------------GACC-CCC ........((((((((((.....((.((((((((..(((.....)))..)))))))).))....((((.....))))))))))..)------------------))).-... ( -35.20) >consensus __GAUUACGUUGUGAAGGAGCACGCCACUCCGGACUCCGUA_______CUCCGGAGUCGCCCAAGUGUUUAAAACACCCUUCAGCC_____C_________UCAGACC_CCC ............((((((.....((.((((((((...............)))))))).))....((((.....))))))))))............................. (-19.73 = -20.73 + 1.00)

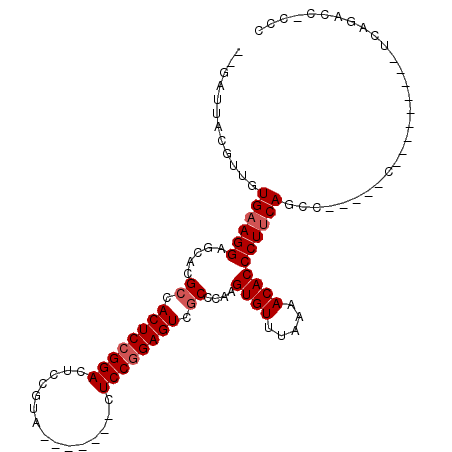
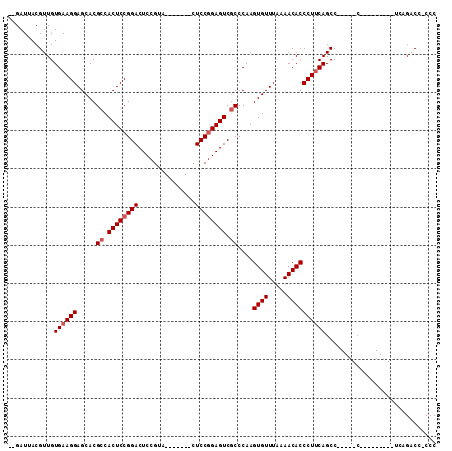
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:49 2006