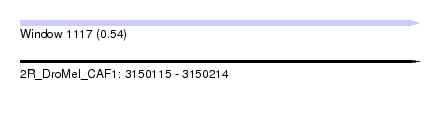
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,150,115 – 3,150,214 |
| Length | 99 |
| Max. P | 0.540530 |

| Location | 3,150,115 – 3,150,214 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 86.00 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
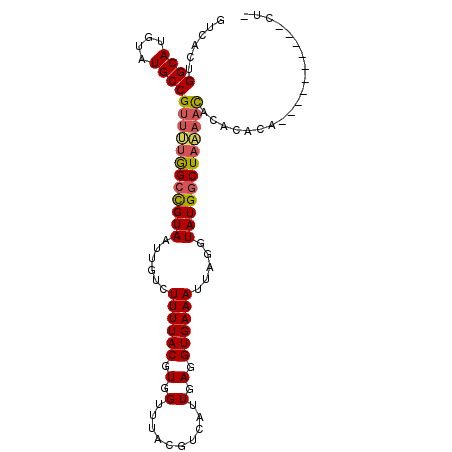
>2R_DroMel_CAF1 3150115 99 + 20766785 GUCACUGGCAUGUAUGCCGUUUUGGCCGUAAUUGUCUUUUACGUGGUUUACGUCAUUGAGGUGAAAUUAGGUAUGGCUAAAACACAACCA----CUCACACUC ......((((....))))((((((((((((.(..((((..(((((...)))))....))))..).......)))))))))))).......----......... ( -25.70) >DroVir_CAF1 49101 94 + 1 AUAACUGGCAUGUAUGCCGUCGUAGCUGUAAUUGUUUUUUACGUUGUUUACGUCAUUGAGGUGAAAUUAGGUAUGCCUUCUCCUCAAUCAGCUC--------- ......((((....)))).....(((((............((((.....)))).(((((((.((....(((....))))).)))))))))))).--------- ( -24.50) >DroSec_CAF1 39879 93 + 1 GUCACUGGCAUGUAUGCCGUUUUGGCCGUAAUUGUGUUUUACGUGGUUUACGUCAUUGAGGUGAAAUUAGGUAUGGCUAAAACACACACA----------UUU ......((((....))))((((((((((((.....(((((((.(.(..........).).)))))))....)))))))))))).......----------... ( -27.20) >DroSim_CAF1 41910 93 + 1 GUCACUGGCAUGUAUGCCGUUUUGGCCGUAAUUGUCUUUUACGUGGUUUACGUCAUUGAGGUGAAAUUAGGUAUGGCUAAAACACACACA----------CUU ......((((....))))((((((((((((.(..((((..(((((...)))))....))))..).......)))))))))))).......----------... ( -25.70) >DroYak_CAF1 39901 87 + 1 GUCACUGGCAUGUAUGCCGUUUUGGCCGUAAUUGUCUUUUACGUGGUUUACGUCAUUGAGGUGAAAUUAGGUAUGGCUAAAACAC----------------UC ......((((....))))((((((((((((.(..((((..(((((...)))))....))))..).......))))))))))))..----------------.. ( -25.70) >DroAna_CAF1 38499 92 + 1 GUCACUGGCAUGUAUGCCGUUGUGGCCGUAAUUGUCUUUUACGUGGUUUACGUCAUUGAGGUGAAAUUAGGUAUGGCUAAAAUACUCACA----------CU- ((((((((((....)))))..))))).(((...(((...(((.((((((((.((...)).))).))))).))).))).....))).....----------..- ( -21.10) >consensus GUCACUGGCAUGUAUGCCGUUUUGGCCGUAAUUGUCUUUUACGUGGUUUACGUCAUUGAGGUGAAAUUAGGUAUGGCUAAAACACACACA__________CU_ ......((((....))))((((((((((((......((((((.(.(..........).).)))))).....)))))))))))).................... (-21.32 = -21.52 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:03 2006