| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,581,638 – 19,581,734 |
| Length | 96 |
| Max. P | 0.993477 |

| Location | 19,581,638 – 19,581,734 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.78 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
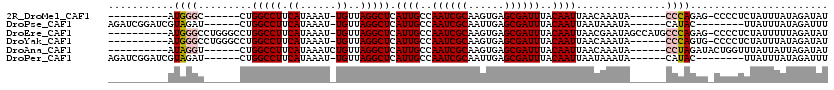
>2R_DroMel_CAF1 19581638 96 + 20766785 ----------AUGGGC------CUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA------CCCAGAG-CCCCUCUAUUUAUAGAUAU ----------..((((------((((........(((-(((..(((.....)))((((((......)))))).))))))........------.)))).)-))).((((....))))... ( -27.83) >DroPse_CAF1 2215 99 + 1 AGAUCGGAUCGUAGAU------CUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAUUGAGCGAUUUACAAUUAAUAAAUA------CAUAC--------UUAUUUAUAGAUUU ((..((((((...)))------)))..)).....(((-(((..(((.....)))((((((......)))))).)))))).(((((((------.....--------.)))))))...... ( -23.00) >DroEre_CAF1 4339 108 + 1 ----------AUGGGCCUGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACGAAUAGCCAUGCCCAGAG-CCCCUCUAUUUUUAGAUAU ----------..((((((((((.((((.(((...(((-(((..(((.....)))((((((......)))))).))))))...)))..)))).)))))).)-))).((((....))))... ( -44.60) >DroYak_CAF1 4338 102 + 1 ----------AUGGGCCUGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA------CCCAGUG-CCCCUCUAUUUAUAGAUAU ----------..(((((((((...(((((.((.....-))..))))).((((..((((((......))))))..)))).........------))))).)-))).((((....))))... ( -32.80) >DroAna_CAF1 4962 98 + 1 ----------AUAGGU------CUGGCCUUCAUAAAUCUGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA------CCUAGAUACUGGUUUAUUAUUAGAUAU ----------.(((((------..(((((.((......))..))))).((((..((((((......))))))..))))........)------))))....(((((.....))))).... ( -20.60) >DroPer_CAF1 5050 99 + 1 AGAUCGGAUCGUAGAU------CUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAUUGAGCGAUUUACAAUUAAUAAAUA------CAUAC--------UUAUUUAUAGAUUU ((..((((((...)))------)))..)).....(((-(((..(((.....)))((((((......)))))).)))))).(((((((------.....--------.)))))))...... ( -23.00) >consensus __________AUAGGC______CUGGCCUUCAUAAAU_UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA______CCCAGAG_CCCCUCUAUUUAUAGAUAU ...........((((.........(((((.((......))..))))).((((..((((((......))))))..))))...............))))....................... (-15.62 = -15.78 + 0.17)
| Location | 19,581,638 – 19,581,734 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -17.99 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
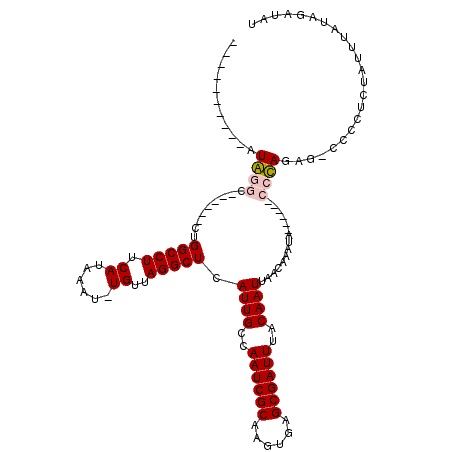
>2R_DroMel_CAF1 19581638 96 - 20766785 AUAUCUAUAAAUAGAGGGG-CUCUGGG------UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAG------GCCCAU---------- ...((((....)))).(((-(.((((.------..((..(.((((((.((((((......))))))(((.....)))..)))-))).)..))..))))------))))..---------- ( -33.70) >DroPse_CAF1 2215 99 - 1 AAAUCUAUAAAUAA--------GUAUG------UAUUUAUUAAUUGUAAAUCGCUCAAUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAG------AUCUACGAUCCGAUCU ..............--------....(------(.(((((.((((((.((((((......))))))(((.....)))..)))-))).))))).)).((------(((........))))) ( -21.90) >DroEre_CAF1 4339 108 - 1 AUAUCUAAAAAUAGAGGGG-CUCUGGGCAUGGCUAUUCGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCAGGCCCAU---------- ...((((....)))).(((-(.((((((.(((((.(((((.((((((.((((((......))))))(((.....)))..)))-))).)))))))))).))))))))))..---------- ( -51.60) >DroYak_CAF1 4338 102 - 1 AUAUCUAUAAAUAGAGGGG-CACUGGG------UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCAGGCCCAU---------- ...((((....)))).(((-(.(((((------(........(((((.((((((......)))))).)))))(.((((....-........)))))..))))))))))..---------- ( -37.60) >DroAna_CAF1 4962 98 - 1 AUAUCUAAUAAUAAACCAGUAUCUAGG------UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACAGAUUUAUGAAGGCCAG------ACCUAU---------- .......................((((------(........(((((.((((((......)))))).)))))(.((((..((......)).)))))..------))))).---------- ( -21.90) >DroPer_CAF1 5050 99 - 1 AAAUCUAUAAAUAA--------GUAUG------UAUUUAUUAAUUGUAAAUCGCUCAAUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAG------AUCUACGAUCCGAUCU ..............--------....(------(.(((((.((((((.((((((......))))))(((.....)))..)))-))).))))).)).((------(((........))))) ( -21.90) >consensus AUAUCUAUAAAUAAAGGGG_CUCUAGG______UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA_AUUUAUGAAGGCCAG______ACCCAU__________ .......................((((...............(((((.((((((......)))))).)))))(.((((.............))))).........))))........... (-17.99 = -17.60 + -0.39)

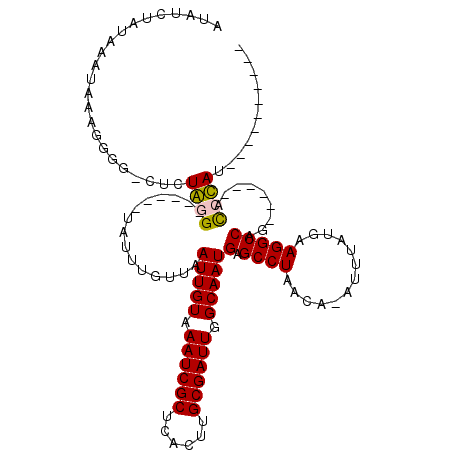
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:15 2006