| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,147,141 – 3,147,289 |
| Length | 148 |
| Max. P | 0.908621 |

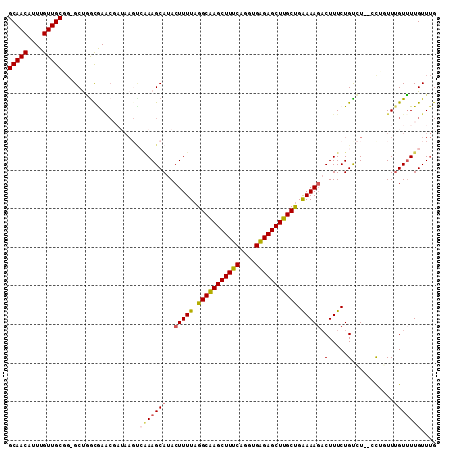
| Location | 3,147,141 – 3,147,249 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -27.49 |
| Energy contribution | -28.27 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
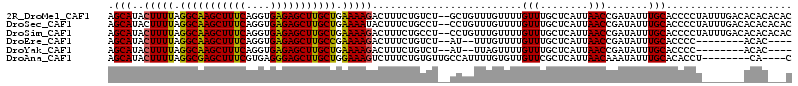
>2R_DroMel_CAF1 3147141 108 + 20766785 GCAACAUUUGUUGCGG-GCUGACAACCGAUAACUCAAAGCAUACUUUUAGGCAAGCUUUCAGGUGAGAGCUUGCUGAAAAGACUUUCUGUCU--GCUGUUUGUUUUGUUUG (((((....)))))((-........)).......(((((((.((((((((.(((((((((....)))))))))))))))((((.....))))--...)).))))))).... ( -36.60) >DroSec_CAF1 36892 108 + 1 GCAACAUUUGUUGCGG-GCUGGCGAACGAUAAGUCAAAGCAUACUUUUAGGCAAGCUUUCAGGUGAGAGCUUGCUGAAAAUACUUUCUGCCU--CCUGUUUGUUUUGUUUG (((((....)))))((-((.(((((((((....))...(((........(((((((((((....)))))))))))((((....)))))))..--...)))))))..)))). ( -33.30) >DroSim_CAF1 37760 106 + 1 GCAACAUUUGUUGCGG-GCUGGCGAACGAUAAGC--AAGCAUACUUUUAGGCAAGCUUUCAGGUGAGAGCUUGCUGAAAAGACUUUCUGCCU--CCUGUUUGUUUUGUUUG (((((....)))))((-((.((((((((....((--(......(((((.(((((((((((....))))))))))).)))))......)))..--..))))))))..)))). ( -37.50) >DroEre_CAF1 36694 106 + 1 GCAACAUUUGUUGCUG-GCUAGUGAACCCGAAGUCGAAGCAUACUUUUAGGCAAGCUUUCAGGUGAGAGCUUGCCGAAAAGACUUUCUGUCU--AU--UUUGUUUUGUUUG (((((....))))).(-(((..((....)).))))((((((........(((((((((((....)))))))))))....((((.....))))--..--..))))))..... ( -34.90) >DroYak_CAF1 37284 106 + 1 GCAACAUUUGUUGCUG-GCCGGUGAGCGAUAAGUCGAAGCAUACUUUUAGGCAAGCUUUCAGGUGAGAGCUUGCUGAAAAGACUUUCUGUCU--AU--UUAGUUUUGUUUG .....((((((((((.-(....).))))))))))((((((.........(((((((((((....)))))))))))(((.((((.....))))--.)--)).)))))).... ( -35.50) >DroAna_CAF1 35863 102 + 1 GCAACAUUUGUUGCUUCGCCCGCAAA---------AAAGCAUACUUUUAGGCGAGCUUUCGUGAGGGAGCUUGCUGGAAAGUCUUUCUGUGUUGCCAUUUUGUGUUGUUCG (((((....)))))......((((((---------(.(((((((((((.((((((((..(....)..)))))))).))))))......)))))....)))))))....... ( -37.80) >consensus GCAACAUUUGUUGCGG_GCUGGCGAACGAUAAGUCAAAGCAUACUUUUAGGCAAGCUUUCAGGUGAGAGCUUGCUGAAAAGACUUUCUGUCU__CCUGUUUGUUUUGUUUG (((((....)))))....................(((((((..(((((.(((((((((((....))))))))))).))))).(.....)...........))))))).... (-27.49 = -28.27 + 0.78)


| Location | 3,147,141 – 3,147,249 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -17.67 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3147141 108 - 20766785 CAAACAAAACAAACAGC--AGACAGAAAGUCUUUUCAGCAAGCUCUCACCUGAAAGCUUGCCUAAAAGUAUGCUUUGAGUUAUCGGUUGUCAGC-CCGCAACAAAUGUUGC ...............((--.(((((((((((((((..(((((((.((....)).)))))))..)))))...)))))((....))..))))).))-..(((((....))))) ( -28.60) >DroSec_CAF1 36892 108 - 1 CAAACAAAACAAACAGG--AGGCAGAAAGUAUUUUCAGCAAGCUCUCACCUGAAAGCUUGCCUAAAAGUAUGCUUUGACUUAUCGUUCGCCAGC-CCGCAACAAAUGUUGC ...............((--.(((..((((((((((..(((((((.((....)).)))))))..)))...)))))))(((.....))).)))...-))(((((....))))) ( -26.80) >DroSim_CAF1 37760 106 - 1 CAAACAAAACAAACAGG--AGGCAGAAAGUCUUUUCAGCAAGCUCUCACCUGAAAGCUUGCCUAAAAGUAUGCUU--GCUUAUCGUUCGCCAGC-CCGCAACAAAUGUUGC .......(((....((.--(((((......(((((..(((((((.((....)).)))))))..)))))..)))))--.))....))).......-..(((((....))))) ( -27.30) >DroEre_CAF1 36694 106 - 1 CAAACAAAACAAA--AU--AGACAGAAAGUCUUUUCGGCAAGCUCUCACCUGAAAGCUUGCCUAAAAGUAUGCUUCGACUUCGGGUUCACUAGC-CAGCAACAAAUGUUGC .............--..--.....(((.(((((((.((((((((.((....)).)))))))).))))).))..))).......(((......))-).(((((....))))) ( -28.70) >DroYak_CAF1 37284 106 - 1 CAAACAAAACUAA--AU--AGACAGAAAGUCUUUUCAGCAAGCUCUCACCUGAAAGCUUGCCUAAAAGUAUGCUUCGACUUAUCGCUCACCGGC-CAGCAACAAAUGUUGC .............--..--.....(((((((......(((((((.((....)).)))))))....(((....))).))))).))(((....)))-..(((((....))))) ( -23.40) >DroAna_CAF1 35863 102 - 1 CGAACAACACAAAAUGGCAACACAGAAAGACUUUCCAGCAAGCUCCCUCACGAAAGCUCGCCUAAAAGUAUGCUUU---------UUUGCGGGCGAAGCAACAAAUGUUGC ((............((....))..((..(((((......))).))..)).))...((((((..(((((....))))---------)..))))))...(((((....))))) ( -23.10) >consensus CAAACAAAACAAACAGG__AGACAGAAAGUCUUUUCAGCAAGCUCUCACCUGAAAGCUUGCCUAAAAGUAUGCUUUGACUUAUCGUUCGCCAGC_CAGCAACAAAUGUUGC ..........................(((((((((..(((((((.((....)).)))))))..)))))...))))......................(((((....))))) (-17.67 = -18.50 + 0.83)

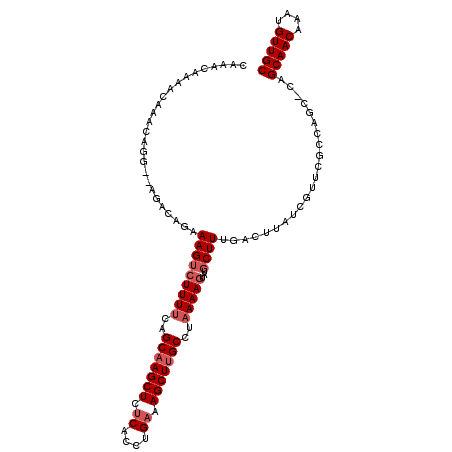
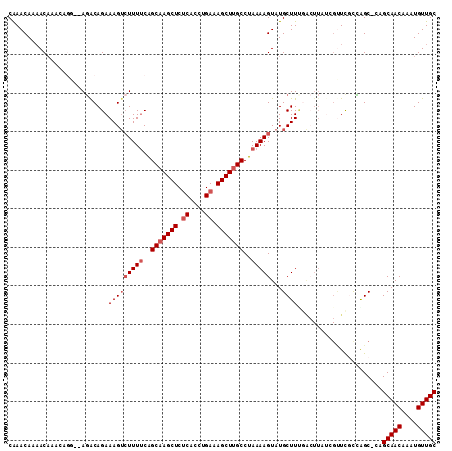
| Location | 3,147,177 – 3,147,289 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.60 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.45 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
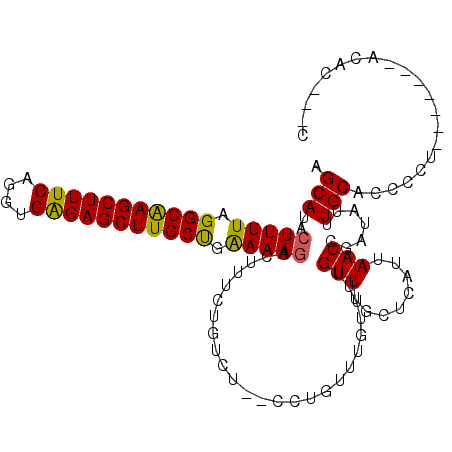
>2R_DroMel_CAF1 3147177 112 + 20766785 AGCAUACUUUUAGGCAAGCUUUCAGGUGAGAGCUUGCUGAAAAGACUUUCUGUCU--GCUGUUUGUUUUGUUUGCUCAUUAACCGAUAUUUGCACCCCUAUUUGACACACACAC ((((...((((.(((((((((((....))))))))))).))))(((.....))))--)))((.((((..((.(((..((........))..))).....))..)))).)).... ( -27.80) >DroSec_CAF1 36928 112 + 1 AGCAUACUUUUAGGCAAGCUUUCAGGUGAGAGCUUGCUGAAAAUACUUUCUGCCU--CCUGUUUGUUUUGUUUGCUCAUUAACCGAUAUUUGCACCCCUAUUUGACACACACAC .(((........(((((((((((....)))))))))))((((....)))))))..--..(((.((((..((.(((..((........))..))).....))..)))).)))... ( -24.00) >DroSim_CAF1 37794 112 + 1 AGCAUACUUUUAGGCAAGCUUUCAGGUGAGAGCUUGCUGAAAAGACUUUCUGCCU--CCUGUUUGUUUUGUUUGCUCAUUAACCGAUAUUUGCACCCCUAUUUGACACACACAC .(((..(((((.(((((((((((....))))))))))).)))))...........--..((((.(((.((......))..))).))))..)))..................... ( -25.80) >DroEre_CAF1 36730 98 + 1 AGCAUACUUUUAGGCAAGCUUUCAGGUGAGAGCUUGCCGAAAAGACUUUCUGUCU--AU--UUUGUUUUGUUUGCUCAUUAACCGAUAUUUGCACCCC--------ACAC---- ((((.((.....(((((((((((....)))))))))))((((((((.....))))--.)--))).....)).))))......................--------....---- ( -28.00) >DroYak_CAF1 37320 98 + 1 AGCAUACUUUUAGGCAAGCUUUCAGGUGAGAGCUUGCUGAAAAGACUUUCUGUCU--AU--UUAGUUUUGUUUGCUCAUUAACCGAUAUUUGCACCCC--------ACAC---- ((((.((.....(((((((((((....)))))))))))(((.((((.....))))--.)--))......)).))))......................--------....---- ( -25.60) >DroAna_CAF1 35891 102 + 1 AGCAUACUUUUAGGCGAGCUUUCGUGAGGGAGCUUGCUGGAAAGUCUUUCUGUGUUGCCAUUUUGUGUUGUUCGCUCAUUAACAAAUAUUUGCACACCU--------CA----C .....((((((.((((((((..(....)..)))))))).)))))).....(((((......(((((..((......))...))))).....)))))...--------..----. ( -32.70) >consensus AGCAUACUUUUAGGCAAGCUUUCAGGUGAGAGCUUGCUGAAAAGACUUUCUGUCU__CCUGUUUGUUUUGUUUGCUCAUUAACCGAUAUUUGCACCCCU_______ACAC___C .(((..(((((.(((((((((((....))))))))))).))))).........................(((........))).......)))..................... (-23.34 = -23.45 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:02 2006