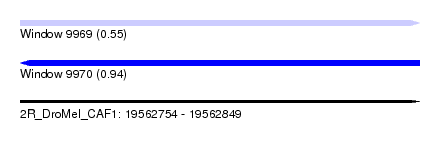
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,562,754 – 19,562,849 |
| Length | 95 |
| Max. P | 0.939534 |

| Location | 19,562,754 – 19,562,849 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -14.20 |
| Energy contribution | -14.64 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
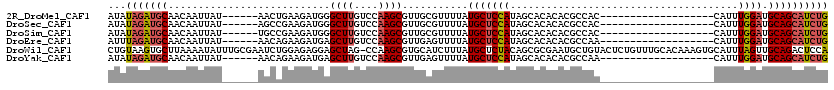
>2R_DroMel_CAF1 19562754 95 + 20766785 AUAUAGAUGCAACAAUUAU------AACUGAAGAUGGGCUUGUCCAAGCGUUGCGUUUUAUGCUCCAUAGCACACACGCCAC-------------------CAUUUGGAUGCAGCAUCUG ...(((((((.........------..((......))((..(((((((.((.((((....((((....))))...)))).))-------------------..))))))))).))))))) ( -24.90) >DroSec_CAF1 2139 95 + 1 AUAUAGAUGCAACAAUUAU------AGCCGAAGAUGGGCUUGUCCAAGCGUUGCGUUUUAUGCUCCAUAGCACACACGCCAC-------------------CAUUUGGAUGCAGCAUCUG ...(((((((.........------((((.......)))).(((((((.((.((((....((((....))))...)))).))-------------------..)))))))...))))))) ( -28.60) >DroSim_CAF1 2107 95 + 1 AUAUAGAUGCAACAAUUAU------UGCCGAAGAUGGGCUUGUCCAAGCGUUGCGUUUUAUGCUCCAUAGCACACACGCCAC-------------------CAUUUGGAUGCAGCAUCUG ...(((((((.........------.(((.......)))..(((((((.((.((((....((((....))))...)))).))-------------------..)))))))...))))))) ( -27.90) >DroEre_CAF1 2076 95 + 1 AUUUAGAUGCAACAAUUAU------AACAGAAGAUGAGCUUGUCCAAGCGUUGAGUUUUAUGCUCCAUAGCACACACGCCAA-------------------CAUUUGGAUGCAGCAUCUG ...(((((((.........------..((.....)).((..(((((((.((((.((....((((....)))).....)))))-------------------).))))))))).))))))) ( -24.80) >DroWil_CAF1 2584 119 + 1 CUGUAAGUGCUUAAAAUAUUUGCGAAUCUGGAGAGGAGCUAG-CCAAGCGUGCAUCUUUAUGCUCUACAGCGCGAAUGCUGUACUCUGUUUGCACAAAGUGCAUUUAGUUGCAGACUCCA ((((((((((......((((((((...(((.(((((......-))..(((((......)))))))).)))))))))))..)))).(((..(((((...)))))..)))))))))...... ( -31.90) >DroYak_CAF1 2241 95 + 1 AUAUAGAUGCAACAAUUAU------AACAGAAGAUGAGCUUGUCCAAGCGUUGAGUUUUAUGCUCCAUAGCACACACGCCAA-------------------CAUUUGGAUGCAGCAUCUG ...(((((((.........------..((.....)).((..(((((((.((((.((....((((....)))).....)))))-------------------).))))))))).))))))) ( -24.90) >consensus AUAUAGAUGCAACAAUUAU______AACAGAAGAUGAGCUUGUCCAAGCGUUGAGUUUUAUGCUCCAUAGCACACACGCCAC___________________CAUUUGGAUGCAGCAUCUG ...(((((((...........................((((....))))...........(((((((......................................)))).)))))))))) (-14.20 = -14.64 + 0.45)
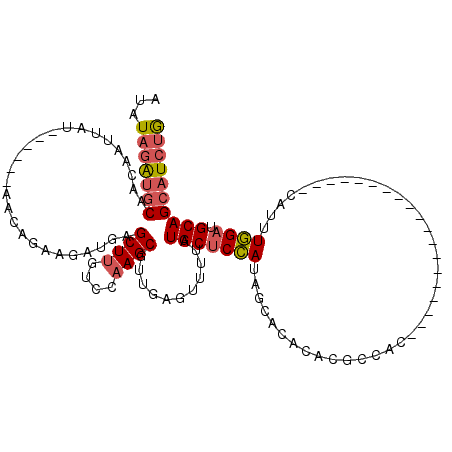
| Location | 19,562,754 – 19,562,849 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -15.98 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19562754 95 - 20766785 CAGAUGCUGCAUCCAAAUG-------------------GUGGCGUGUGUGCUAUGGAGCAUAAAACGCAACGCUUGGACAAGCCCAUCUUCAGUU------AUAAUUGUUGCAUCUAUAU .((((((.((((((((.((-------------------...((((.((((((....))))))..))))..)).)))))..(((.........)))------.....))).)))))).... ( -27.40) >DroSec_CAF1 2139 95 - 1 CAGAUGCUGCAUCCAAAUG-------------------GUGGCGUGUGUGCUAUGGAGCAUAAAACGCAACGCUUGGACAAGCCCAUCUUCGGCU------AUAAUUGUUGCAUCUAUAU .((((((.((((((((.((-------------------...((((.((((((....))))))..))))..)).)))))..((((.......))))------.....))).)))))).... ( -33.50) >DroSim_CAF1 2107 95 - 1 CAGAUGCUGCAUCCAAAUG-------------------GUGGCGUGUGUGCUAUGGAGCAUAAAACGCAACGCUUGGACAAGCCCAUCUUCGGCA------AUAAUUGUUGCAUCUAUAU .((((((.((((((((.((-------------------...((((.((((((....))))))..))))..)).)))))...(((.......))).------.....))).)))))).... ( -32.80) >DroEre_CAF1 2076 95 - 1 CAGAUGCUGCAUCCAAAUG-------------------UUGGCGUGUGUGCUAUGGAGCAUAAAACUCAACGCUUGGACAAGCUCAUCUUCUGUU------AUAAUUGUUGCAUCUAAAU .((((((.((((((((.((-------------------((((....((((((....))))))....)))))).)))))..(((.........)))------.....))).)))))).... ( -27.80) >DroWil_CAF1 2584 119 - 1 UGGAGUCUGCAACUAAAUGCACUUUGUGCAAACAGAGUACAGCAUUCGCGCUGUAGAGCAUAAAGAUGCACGCUUGG-CUAGCUCCUCUCCAGAUUCGCAAAUAUUUUAAGCACUUACAG (((((..((((.((..(((((((((((....)))))))(((((......)))))...))))..)).)))).(((...-..)))....))))).....((...........))........ ( -31.70) >DroYak_CAF1 2241 95 - 1 CAGAUGCUGCAUCCAAAUG-------------------UUGGCGUGUGUGCUAUGGAGCAUAAAACUCAACGCUUGGACAAGCUCAUCUUCUGUU------AUAAUUGUUGCAUCUAUAU .((((((.((((((((.((-------------------((((....((((((....))))))....)))))).)))))..(((.........)))------.....))).)))))).... ( -27.80) >consensus CAGAUGCUGCAUCCAAAUG___________________GUGGCGUGUGUGCUAUGGAGCAUAAAACGCAACGCUUGGACAAGCCCAUCUUCAGUU______AUAAUUGUUGCAUCUAUAU .((((((.(((.............................(((((.((((((....)))))).......)))))(((......)))....................))).)))))).... (-15.98 = -16.82 + 0.84)

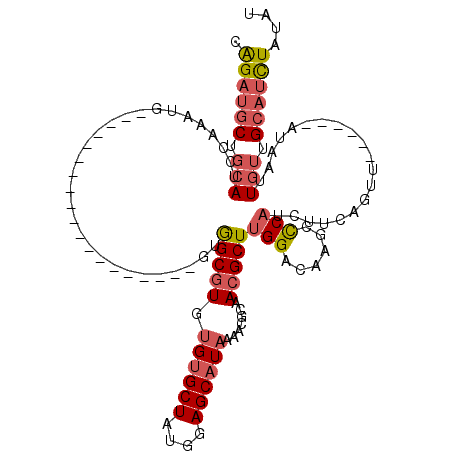

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:02 2006