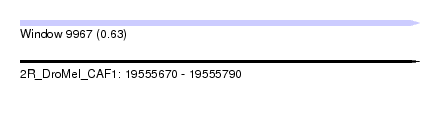
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,555,670 – 19,555,790 |
| Length | 120 |
| Max. P | 0.630199 |

| Location | 19,555,670 – 19,555,790 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.89 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -31.68 |
| Energy contribution | -31.49 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19555670 120 + 20766785 UGACUUCUAGAGCAUGCUGAUAGAAGCGCAAAGCCAUGUCAGGAUGCUUGGAUUCAGUUAACUUGGCGGCUUUAUCCAAGGCGGAUGCGGCUGCCUCUGGGGAACCGUGCUGUUGAUACA (((..((((.(((((.((((((...((.....))..)))))).))))))))).)))((((((..(((((((.(((((.....))))).)))))))...((....)).....))))))... ( -47.50) >DroAna_CAF1 2935 120 + 1 UAACUUCUAGAGCAUGUUGAUAGAAUCGCAGUGCCAUGUCAGGGUGCUUAUUUUCGGUUAGUUUGGCAGCUUUGUCCAGAGCAGUGGCUGCUGCCUCCGGAGAACCAUGCUGCUGAUAAA (((((....((((((.((((((.....((...))..)))))).))))))......)))))..(..(((((.((.(((.(((((((....)))).))).))).))....)))))..).... ( -38.50) >DroSim_CAF1 2662 120 + 1 UGACUUCUAGAGCAUGCUGAUAGAAGCGCAAAGCCAUGUCAGGAUGCUUGGAUUCAGUUAACUUGGCGGCUUUAUCCAAGGCGGAUGCUGCUGCCUCUGGGGAACCGUGCUGUUGAUACA (((..((((.(((((.((((((...((.....))..)))))).))))))))).)))((((((..((((((..(((((.....)))))..))))))...((....)).....))))))... ( -46.20) >DroEre_CAF1 2847 120 + 1 UAACUUCUAAAGCAUGCUGAUAAAACCGCAAAGCCAUGUCAGGAUGCUUGGAUUCAGUUAACUUUGCGGCUUUAUCCAAGGCGGAUGCGGCUGCCUCUGGGGAACCAUGCUGUUGAUACA .....(((((.((((.((((((..............)))))).)))))))))....((((((...((((((.(((((.....))))).))))))...(((....)))....))))))... ( -38.04) >DroWil_CAF1 2719 120 + 1 UCACUUCGCCAGCAUGUUGAUAGAAUCGAAGAGCCAUUUCUGGAUGCUUUUGCUCAGUUAGUUUUGCAGCUUUGUCCAAUGCGGAUGCUGCAGCUUCAGGAGAUCCAUGUUGCUGAUACA .....(((.(((((((..(((.......((((((.(((....))))))))).(((....(((..((((((...((((.....)))))))))))))....))))))))))))).))).... ( -32.60) >DroYak_CAF1 2749 120 + 1 UAACUUCUAGAGCAUGCUGAUAGAACCGCAAGGCCAUGUCAGGAUGCUUGGAUUCAGUUAACUUAGCGGCUUUAUCCAAGGCGGAUGCAGCUGCCUCGGGGGAACCAUGCUGUUGAUACA (((((((((.(((((.((((((...((....))...)))))).)))))))))...)))))..((((((((......(.((((((......)))))).)((....))..)))))))).... ( -46.30) >consensus UAACUUCUAGAGCAUGCUGAUAGAACCGCAAAGCCAUGUCAGGAUGCUUGGAUUCAGUUAACUUGGCGGCUUUAUCCAAGGCGGAUGCUGCUGCCUCUGGGGAACCAUGCUGUUGAUACA (((((....((((((.((((((..............)))))).))))))......)))))..((((((((........((((((......))))))..((....))..)))))))).... (-31.68 = -31.49 + -0.19)

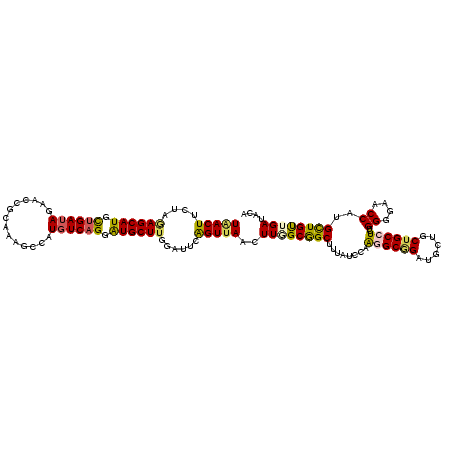
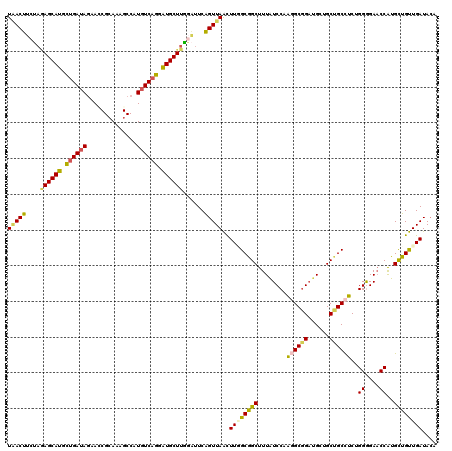
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:59 2006