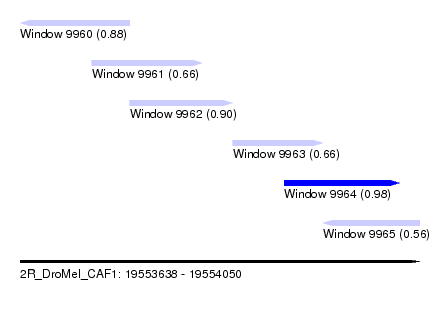
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,553,638 – 19,554,050 |
| Length | 412 |
| Max. P | 0.976677 |

| Location | 19,553,638 – 19,553,751 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.40 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -18.72 |
| Energy contribution | -20.76 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19553638 113 - 20766785 UU-UUUUAUAUUUCGAUGGCUAUAAAUAUGUGCGUAUGUAUGUAUCUGCGGGGCAAUGCUUAAUGCUUAUUUUCUC-----GCAGACGCAAAAUGUGUGUACAUAUGUACGUCCAUAU-U ..-.....................((((((..(((((((((((((((((((((.(((((.....))..))).))))-----))))((((.....)))))))))))))))))..)))))-) ( -37.20) >DroSec_CAF1 781 114 - 1 UUUUUUUAUAUUUCGAUGGCUAUAAAUAUGUGCAUAUGUAUGUAUCUGCGGGGCAAUGCUUAAUACUUAUUUUCUC-----GCAGACGCAAAAUGUGUGUACAUAUGUACGUCCAUAU-U ...............((((......((((((((((((((.((..(((((((((.((((.........)))).))))-----)))))..))..))))))))))))))......))))..-. ( -35.20) >DroAna_CAF1 812 105 - 1 UU-UU---UAUUCCGAUGGCUAUAAAUAUGUACAUAUGUAUGUAUCUGCGGGGAAAUGCUU-------AUGUUCUCCUAUGCCAGACGCAAACUGUGUGU----AUGUAAAAAUAUAAAA ..-..---..................(((((((((((((.((..((((((((((...((..-------..))..)))).)).))))..)).)).))))))----)))))........... ( -23.30) >DroSim_CAF1 642 113 - 1 UU-UUUUAUAUUUCGAUGGCUAUAAAUAUGUGCGUAUGUAUGUAUCUGCGGGGCAAUGCUUAAUGCUUAUUUUCUC-----GCAGACGCAAAAUGUGUGUACAUAUGUACGUCCAUAU-U ..-.....................((((((..(((((((((((((((((((((.(((((.....))..))).))))-----))))((((.....)))))))))))))))))..)))))-) ( -37.20) >DroYak_CAF1 663 109 - 1 UU-UUUUAUAAUUCGAUGGCUAUAAAUAUGUGCGUAUGUAUGUAUCUGCGGGGCAAUGCUUAAUGCUUAUUUUCUC-----GCAGACGCAAAAUGUGUGU----AUGUACGUCCAUAU-U ..-.....................((((((..(((((((((....((((((((.(((((.....))..))).))))-----))))((((.....))))))----)))))))..)))))-) ( -31.10) >consensus UU_UUUUAUAUUUCGAUGGCUAUAAAUAUGUGCGUAUGUAUGUAUCUGCGGGGCAAUGCUUAAUGCUUAUUUUCUC_____GCAGACGCAAAAUGUGUGUACAUAUGUACGUCCAUAU_U ...............((((......((((((((((((((.((..(((((((((.((((.........)))).)))).....)))))..))..))))))))))))))......)))).... (-18.72 = -20.76 + 2.04)

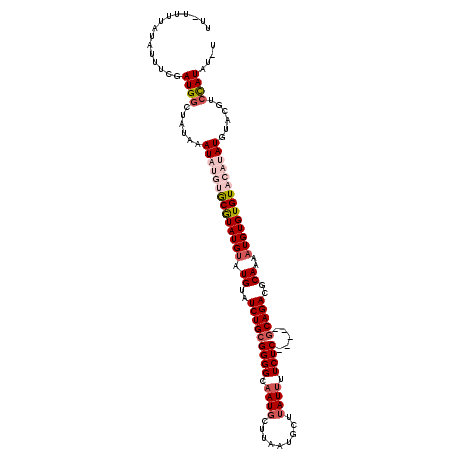
| Location | 19,553,712 – 19,553,826 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -18.84 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19553712 114 + 20766785 UACAUACGCACAUAUUUAUAGCCAUCGAAAUAUAAAA-AAAGCGGUAUACCCACACACGCUCAAGUGUGUAAAAAUAUAUAUAUGUAUAGACA----UGAAAAUCCA-AUGUGUGUGCAU ....(((((((...((((((..........)))))).-..((((((........)).))))...)))))))...........(((((((.(((----(.........-)))).))))))) ( -24.10) >DroSec_CAF1 855 115 + 1 UACAUAUGCACAUAUUUAUAGCCAUCGAAAUAUAAAAAAAAGCGGUAUACCCACACACGCUCAAGUGUGUAAAAAUAUAUAUAUGUAUAGACA----UGAAAAUCCA-AUGUGUGUGCAU .....((((((((((..............(((((.......(.((....)))((((((......)))))).....))))).(((((....)))----))........-..)))))))))) ( -24.10) >DroAna_CAF1 881 116 + 1 UACAUAUGUACAUAUUUAUAGCCAUCGGAAUA---AA-AAAGCGGUAUACCCACACACACUCAUGUGUGUAAAAAUAUAUGUAUACAUAUAUACCCCACAAAAUGCAUAUGUGUGCGCAU ((((((((((....(((((..((...)).)))---))-...(.((((.....(((((((....))))))).....((((((....)))))))))).)......))))))))))....... ( -25.70) >DroSim_CAF1 716 114 + 1 UACAUACGCACAUAUUUAUAGCCAUCGAAAUAUAAAA-AAAGCGGUAUACCCACACACGCUCAAGUGUGUAAAAAUAUAUAUAUGUAUAGACA----UGAAAAUCCA-AUGUGUGUGCAU ....(((((((...((((((..........)))))).-..((((((........)).))))...)))))))...........(((((((.(((----(.........-)))).))))))) ( -24.10) >DroEre_CAF1 888 114 + 1 UACAUACGCACAUAUUUAUAGCCAUCGAAUUAUAAAA-AAAGCGGUAUACCCACACACGCUCAAGUGUGUAAAAAUAUAUAUAUGUAUAGACA----UGAAAACCCA-AUGUGUGUGCAU ....(((((((((.(((((((........))))))).-...(.(((......((((((......))))))...........(((((....)))----))...)))).-)))))))))... ( -24.90) >DroYak_CAF1 733 114 + 1 UACAUACGCACAUAUUUAUAGCCAUCGAAUUAUAAAA-AAAGCGGUAUACCCACACACGCUCAAGUGUGUAAAAAUAUAUAUAUGUAUAGACA----UGAAAAUCCA-AUGUGUGUGCAU ....(((((((...(((((((........))))))).-..((((((........)).))))...)))))))...........(((((((.(((----(.........-)))).))))))) ( -24.60) >consensus UACAUACGCACAUAUUUAUAGCCAUCGAAAUAUAAAA_AAAGCGGUAUACCCACACACGCUCAAGUGUGUAAAAAUAUAUAUAUGUAUAGACA____UGAAAAUCCA_AUGUGUGUGCAU .......((((((((..........................(.((....)))((((((......))))))........................................)))))))).. (-18.84 = -18.87 + 0.03)
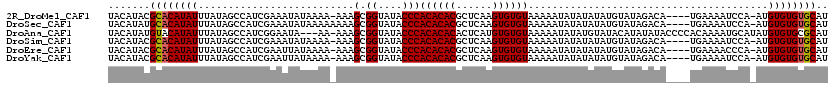
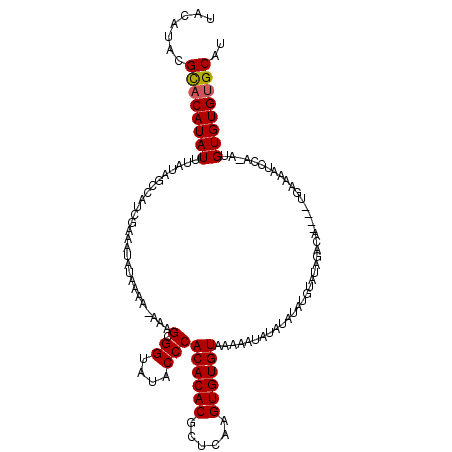
| Location | 19,553,751 – 19,553,857 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 91.82 |
| Mean single sequence MFE | -22.84 |
| Consensus MFE | -18.83 |
| Energy contribution | -19.22 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19553751 106 + 20766785 AGCGGUAUACCCACACACGCUCAAGUGUGUAAAAAUAUAUAUAUGUAUAGACA----UGAAAAUCCA-AUGUGUGUGCAUUUCUCUUUCCAACAUUUCAAUUUUUAAUCGC------- .(((((......((((((......))))))............(((((((.(((----(.........-)))).)))))))..........................)))))------- ( -22.60) >DroSec_CAF1 895 106 + 1 AGCGGUAUACCCACACACGCUCAAGUGUGUAAAAAUAUAUAUAUGUAUAGACA----UGAAAAUCCA-AUGUGUGUGCAUUUCUCUUUCCAACAUUUCAAUUUUUAAUCGC------- .(((((......((((((......))))))............(((((((.(((----(.........-)))).)))))))..........................)))))------- ( -22.60) >DroAna_CAF1 917 118 + 1 AGCGGUAUACCCACACACACUCAUGUGUGUAAAAAUAUAUGUAUACAUAUAUACCCCACAAAAUGCAUAUGUGUGCGCAUUUCUCUUUCCAACUUUUUAUUUUUUAAUCGCACCAGCG .((((((.....(((((((....))))))).....((((((....))))))))))..............((.(((((.(((........................)))))))))))). ( -24.06) >DroSim_CAF1 755 106 + 1 AGCGGUAUACCCACACACGCUCAAGUGUGUAAAAAUAUAUAUAUGUAUAGACA----UGAAAAUCCA-AUGUGUGUGCAUUUCUCUUUCCAACAUUUCAAUUUUUAAUCGC------- .(((((......((((((......))))))............(((((((.(((----(.........-)))).)))))))..........................)))))------- ( -22.60) >DroEre_CAF1 927 106 + 1 AGCGGUAUACCCACACACGCUCAAGUGUGUAAAAAUAUAUAUAUGUAUAGACA----UGAAAACCCA-AUGUGUGUGCAUUUCUCUUUCCAACAUUUCAAUUUUUAAUCGC------- .(((((......((((((......))))))............(((((((.(((----(.........-)))).)))))))..........................)))))------- ( -22.60) >DroYak_CAF1 772 106 + 1 AGCGGUAUACCCACACACGCUCAAGUGUGUAAAAAUAUAUAUAUGUAUAGACA----UGAAAAUCCA-AUGUGUGUGCAUUUCUCUUUCCAACAUUUCAAUUUUUAAUCGC------- .(((((......((((((......))))))............(((((((.(((----(.........-)))).)))))))..........................)))))------- ( -22.60) >consensus AGCGGUAUACCCACACACGCUCAAGUGUGUAAAAAUAUAUAUAUGUAUAGACA____UGAAAAUCCA_AUGUGUGUGCAUUUCUCUUUCCAACAUUUCAAUUUUUAAUCGC_______ .(((((......((((((......))))))............(((((((.(((....((......))..))).)))))))..........................)))))....... (-18.83 = -19.22 + 0.39)
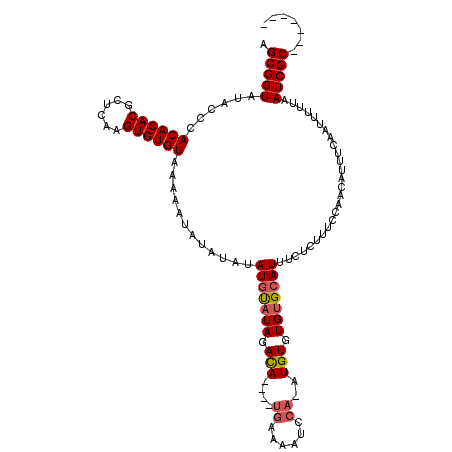
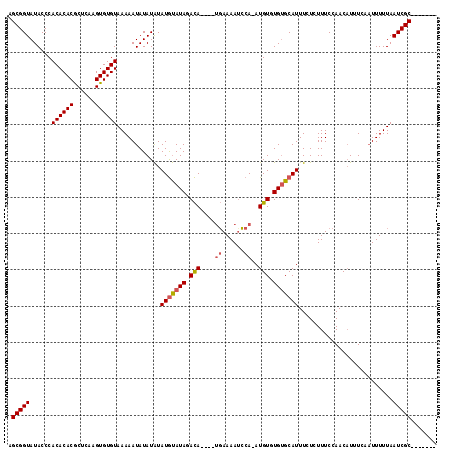
| Location | 19,553,857 – 19,553,950 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 84.25 |
| Mean single sequence MFE | -33.41 |
| Consensus MFE | -19.43 |
| Energy contribution | -21.49 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19553857 93 + 20766785 -UCCAUUUUUCCAGCUGCCACUUUGGA---GCAUUCUUUC----GCGAUUGCCUUUCGCGAAUUUGCCAGUUAUGAGUGUGAGUGGCGGCUGAGAAGUAUG -..(((.((((((((((((((((....---((((((.(((----((((.......)))))))............)))))))))))))))))).)))).))) ( -39.12) >DroSec_CAF1 1001 93 + 1 -UCCAUUUUUCCAGCUGCCACUUUGGA---GCAUUCUUUC----GCGAUUGCCUUUCGCGAAUUUGCCAGUUAUGAGUGUGAGUGGCGGCUGAGAAGUAUG -..(((.((((((((((((((((....---((((((.(((----((((.......)))))))............)))))))))))))))))).)))).))) ( -39.12) >DroAna_CAF1 1035 93 + 1 AUCCAUUUUUCCAGAUGUUACAAUAAAUUCGCACUCUUGCGUCGGCGAUUGGUUUU--------AACCAUUAAUGAGUGUGAGUGAUUCGCGAAAAGUAUG ....(((((((..((.(((((.......((((((((((((....)))).((((...--------.)))).....)))))))))))))))..)))))))... ( -22.71) >DroSim_CAF1 861 93 + 1 -UCCAUUUUUCCAGCUGCCACUUUGGA---GCAUUCUUUC----GCGAUUGCCUUUCGCGAAUUUGCCAGUUAUGAGUGUGAGUGGCGGCUGAGAAGUAUG -..(((.((((((((((((((((....---((((((.(((----((((.......)))))))............)))))))))))))))))).)))).))) ( -39.12) >DroEre_CAF1 1033 93 + 1 -UCCAUUUUUCCAGCUGCCACUUUUGA---GCAUUCUUUC----GCGAUUGCCUUUCGCGAAUUUGCCAGUUAUGAGUGUGAGCGGCGACGGAGAUGUAUG -..(((.(((((.(((((((((((((.---(((....(((----((((.......)))))))..))))))....)))))...)))))...)))))...))) ( -30.80) >DroYak_CAF1 878 93 + 1 -UCCAUUUUUCCAGCUGCCACUUUUGA---GCAUUCUUUC----GCGAUUGCCUUUCGCGAAUUUGCCAUUUAUGAGUGUGAGUGUCGGCGGAGAUGUAUG -..(((.(((((.((((.(((((....---((((((.(((----((((.......)))))))............))))))))))).)))))))))...))) ( -29.62) >consensus _UCCAUUUUUCCAGCUGCCACUUUGGA___GCAUUCUUUC____GCGAUUGCCUUUCGCGAAUUUGCCAGUUAUGAGUGUGAGUGGCGGCUGAGAAGUAUG ....(((((((..((((((((((.......((((((........((((((((.....))))..)))).......)))))))))))))))).)))))))... (-19.43 = -21.49 + 2.06)

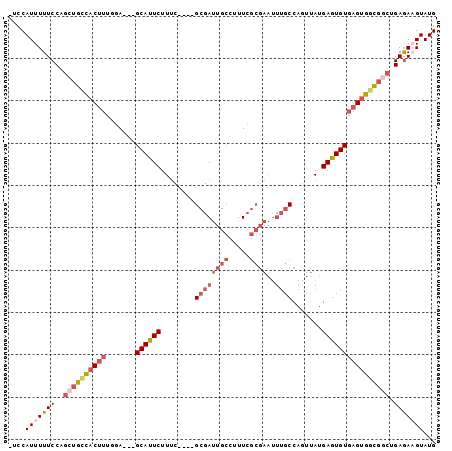
| Location | 19,553,910 – 19,554,029 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.40 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -28.96 |
| Energy contribution | -30.30 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19553910 119 + 20766785 AUUUGCCAGUUAUGAGUGUGAGUGGCGGCUGAGAAGUAUG-AAACACUCCAAUAAAUGAGUAAUAUCCGCUGCUACCAAUGACUAAUUUGGUAUAUAUGUAUACGCACACAUUCAUGACC ........((((((((((((.((((((((.((...((...-..))((((........))))....)).)))))((((((........))))))..........))).)))))))))))). ( -36.60) >DroSec_CAF1 1054 119 + 1 AUUUGCCAGUUAUGAGUGUGAGUGGCGGCUGAGAAGUAUG-AAACACUCCAAUAAAUGAGUAAUAUCCGCUGCUACCAAUGACUAAUUUGGUAUAUAUGUAUACGCACACAUUCAUGACC ........((((((((((((.((((((((.((...((...-..))((((........))))....)).)))))((((((........))))))..........))).)))))))))))). ( -36.60) >DroAna_CAF1 1091 117 + 1 ---AACCAUUAAUGAGUGUGAGUGAUUCGCGAAAAGUAUGGAAACACUUCAGUAAAUGAGUAAUAUCCGUUGCUACCAAUGACUAAUUUGGUUUAUAUGUAUACGCACACAUCCAUGGCU ---..((((..(((.(((((.(((....((((...(((((....)(((.((.....))))).))))...)))).(((((........))))).......))).))))).)))..)))).. ( -24.60) >DroSim_CAF1 914 119 + 1 AUUUGCCAGUUAUGAGUGUGAGUGGCGGCUGAGAAGUAUG-GAACACUCCAAUAAAUGAGUAAUAUCCGCUGCUACCAAUGACUAAUUUGGUAUAUAUGUAUACGCACACAUUCAUGACC ........((((((((((((.((((((((.((...((...-..))((((........))))....)).)))))((((((........))))))..........))).)))))))))))). ( -36.60) >DroEre_CAF1 1086 119 + 1 AUUUGCCAGUUAUGAGUGUGAGCGGCGACGGAGAUGUAUG-AAACACUCCAAUAAAUGAGUAAUAUCCGCUGCUACCAAUGACUAAUUUGGUAUAUAUGUAUACGCACACACUCAUGACC ........(((((((((((((((((((..((((.(((...-..))))))).......((......)))))))))(((((........)))))...............)))))))))))). ( -41.70) >DroYak_CAF1 931 119 + 1 AUUUGCCAUUUAUGAGUGUGAGUGUCGGCGGAGAUGUAUG-AAACACUCCAAUAAAUGAGUAAUAUCCGCUGCUACCAAUGACUAAUUUGGUAUAUAUGUAUACGCACACAAUCAUGACC .........((((((.((((.(((((((((((...((...-..))((((........))))....))))))).((((((........)))))).........)))).)))).)))))).. ( -31.90) >consensus AUUUGCCAGUUAUGAGUGUGAGUGGCGGCUGAGAAGUAUG_AAACACUCCAAUAAAUGAGUAAUAUCCGCUGCUACCAAUGACUAAUUUGGUAUAUAUGUAUACGCACACAUUCAUGACC ........((((((((((((.((((((((.((...((......))((((........))))....)).)))))((((((........))))))..........))).)))))))))))). (-28.96 = -30.30 + 1.34)

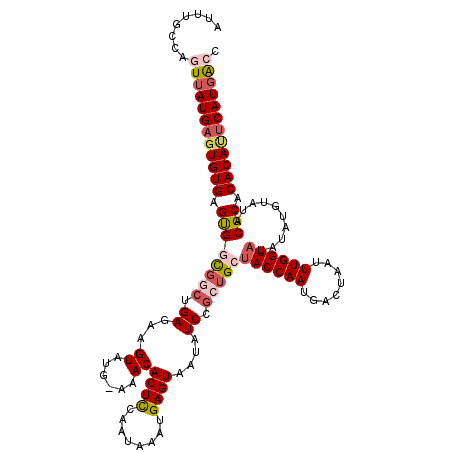
| Location | 19,553,950 – 19,554,050 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 94.82 |
| Mean single sequence MFE | -22.19 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.67 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19553950 100 - 20766785 GCCAUAUUCGGGCAUAUA----CAUGGUCAUGAAUGUGUGCGUAUACAUAUAUACCAAAUUAGUCAUUGGUAGCAGCGGAUAUUACUCAUUUAUUGGAGUGUUU- ...(((((((.((.((((----(((........)))))))............((((((........))))))))..)))))))(((((........)))))...- ( -22.30) >DroSec_CAF1 1094 104 - 1 GCCAUAUUCGGGCAUAUAAAUAAAUGGUCAUGAAUGUGUGCGUAUACAUAUAUACCAAAUUAGUCAUUGGUAGCAGCGGAUAUUACUCAUUUAUUGGAGUGUUU- (((((((((((((.(((......)))))).)))))))).))((((.......((((((........)))))).......))))(((((........)))))...- ( -22.64) >DroAna_CAF1 1128 101 - 1 GCCAUAUUCGGGCAUAUA----CAUAGCCAUGGAUGUGUGCGUAUACAUAUAAACCAAAUUAGUCAUUGGUAGCAACGGAUAUUACUCAUUUACUGAAGUGUUUC (((((((((((((.....----....))).)))))))).))............(((((........)))))......(((((((..(((.....)))))))))). ( -19.70) >DroSim_CAF1 954 100 - 1 GCCAUAUUCGGGCAUAUA----CAUGGUCAUGAAUGUGUGCGUAUACAUAUAUACCAAAUUAGUCAUUGGUAGCAGCGGAUAUUACUCAUUUAUUGGAGUGUUC- ...(((((((.((.((((----(((........)))))))............((((((........))))))))..)))))))(((((........)))))...- ( -22.30) >DroEre_CAF1 1126 100 - 1 GCCAUAUUCGGGCAUAUA----CAUGGUCAUGAGUGUGUGCGUAUACAUAUAUACCAAAUUAGUCAUUGGUAGCAGCGGAUAUUACUCAUUUAUUGGAGUGUUU- ...(((((((.(((((((----(..........))))))))...........((((((........))))))....)))))))(((((........)))))...- ( -23.60) >DroYak_CAF1 971 100 - 1 GCCAUAUUCGGGCAUAUA----CAUGGUCAUGAUUGUGUGCGUAUACAUAUAUACCAAAUUAGUCAUUGGUAGCAGCGGAUAUUACUCAUUUAUUGGAGUGUUU- ...(((((((.(((..((----((..((....))..))))..).........((((((........))))))))..)))))))(((((........)))))...- ( -22.60) >consensus GCCAUAUUCGGGCAUAUA____CAUGGUCAUGAAUGUGUGCGUAUACAUAUAUACCAAAUUAGUCAUUGGUAGCAGCGGAUAUUACUCAUUUAUUGGAGUGUUU_ (((((((((((((.(((......)))))).)))))))).))((((.......((((((........)))))).......))))(((((........))))).... (-21.70 = -21.67 + -0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:56 2006