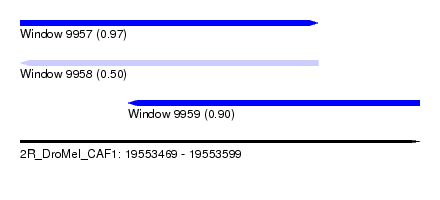
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,553,469 – 19,553,599 |
| Length | 130 |
| Max. P | 0.968506 |

| Location | 19,553,469 – 19,553,566 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 90.19 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -20.94 |
| Energy contribution | -23.34 |
| Covariance contribution | 2.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
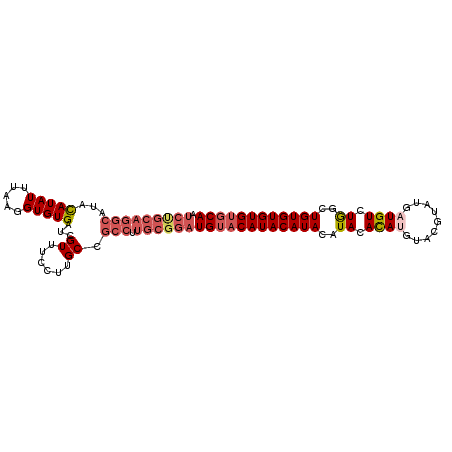
>2R_DroMel_CAF1 19553469 97 + 20766785 AAUCAAAAUCGUCCCACAUGCAAACAGUCGUCUGCUGCACACACACAGCUAAACAU--------CAUGUGUAUGUAUGUAUGAACAUCCGCAAGGCGGCAAGGAA ...........(((....((....))(((((((((((........)))).......--------..((((.((((........)))).)))))))))))..))). ( -21.20) >DroSec_CAF1 608 101 + 1 AAUCAAAAUCGUCCCACAUGC----AGCCGUCUGCUGCACACACACAGCCAGACAUAAUACGUACAUGUGUAUGUAUGUAUGUACAUCCGCAAGGCGGCAAGGAA ...........(((....(((----(((.....))))))........(((..(((((((((((((....)))))))).)))))....((....)).)))..))). ( -32.10) >DroSim_CAF1 477 105 + 1 AAUCAAAAUCGUCCCACAUGCAAACAGCCGUCUGCUGCACACACACAGCCAGACAAAAUACGUACAUGUGUAUGUAUGUAUGUACAUCCGCAAGGCGGCAAGGAA ...........(((.((((((........((((((((........))).)))))...((((((((....))))))))))))))....((((...))))...))). ( -30.70) >DroEre_CAF1 655 105 + 1 AAUCAAAAUCGUCACACAUGCAUACAGCCGUCUGCUGCACACACACAUCCAGACAUCAUACGUACAUGUGUAUGUAUGUAUGUACAUCCGCAAGGCGGCAAGGAA ..........(((..(((((((((((...(((((.((........))..)))))...(((((....))))).)))))))))))....((....)).)))...... ( -27.90) >DroYak_CAF1 487 104 + 1 AAUCAAAAUCGUCACACAUGCAAACAGCCGUCUGCUG-ACACACACAACCAGACAUCACACGUACUUAUGUAUGUAUGUAUGUACAUCCGCAAGGCGGCAAGGAA ..........(((..(((((((.......(((((.((-.......))..))))).....((((((....)))))).)))))))....((....)).)))...... ( -24.80) >consensus AAUCAAAAUCGUCCCACAUGCAAACAGCCGUCUGCUGCACACACACAGCCAGACAUAAUACGUACAUGUGUAUGUAUGUAUGUACAUCCGCAAGGCGGCAAGGAA ..........(((..((((((........((((((((........)))).))))...((((((((....))))))))))))))....((....)).)))...... (-20.94 = -23.34 + 2.40)

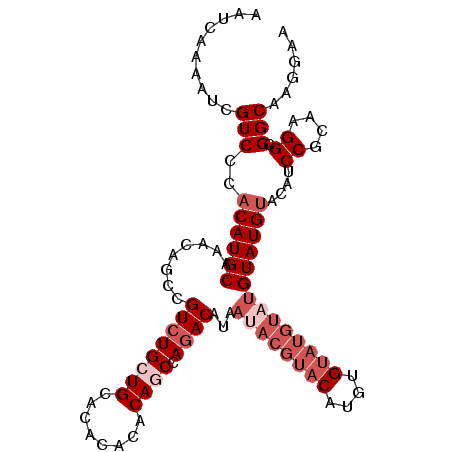
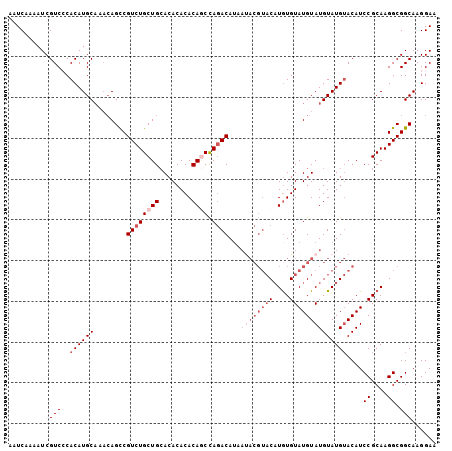
| Location | 19,553,469 – 19,553,566 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.19 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -23.12 |
| Energy contribution | -24.08 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19553469 97 - 20766785 UUCCUUGCCGCCUUGCGGAUGUUCAUACAUACAUACACAUG--------AUGUUUAGCUGUGUGUGUGCAGCAGACGACUGUUUGCAUGUGGGACGAUUUUGAUU .......((((...)))).(((((.((((((((((((((.(--------........))))))))))...((((((....))))))))))))))))......... ( -29.00) >DroSec_CAF1 608 101 - 1 UUCCUUGCCGCCUUGCGGAUGUACAUACAUACAUACACAUGUACGUAUUAUGUCUGGCUGUGUGUGUGCAGCAGACGGCU----GCAUGUGGGACGAUUUUGAUU ((((.(((.(((...((((((((.((((.(((((....))))).))))))))))))(((((......)))))....))).----)))...))))........... ( -33.10) >DroSim_CAF1 477 105 - 1 UUCCUUGCCGCCUUGCGGAUGUACAUACAUACAUACACAUGUACGUAUUUUGUCUGGCUGUGUGUGUGCAGCAGACGGCUGUUUGCAUGUGGGACGAUUUUGAUU .(((...((((...))))(((((......))))).((((((((..((..(((((((.((((......))))))))))).))..)))))))))))........... ( -32.30) >DroEre_CAF1 655 105 - 1 UUCCUUGCCGCCUUGCGGAUGUACAUACAUACAUACACAUGUACGUAUGAUGUCUGGAUGUGUGUGUGCAGCAGACGGCUGUAUGCAUGUGUGACGAUUUUGAUU ....(((.(((....(((((((.(((((.(((((....))))).))))))))))))...(..((((((((((.....))))))))))..)))).)))........ ( -33.20) >DroYak_CAF1 487 104 - 1 UUCCUUGCCGCCUUGCGGAUGUACAUACAUACAUACAUAAGUACGUGUGAUGUCUGGUUGUGUGUGU-CAGCAGACGGCUGUUUGCAUGUGUGACGAUUUUGAUU .......((((...)))).(((.(((((((((((((((((.((.(........))).))))))))))-..((((((....))))))))))))))))......... ( -30.00) >consensus UUCCUUGCCGCCUUGCGGAUGUACAUACAUACAUACACAUGUACGUAUGAUGUCUGGCUGUGUGUGUGCAGCAGACGGCUGUUUGCAUGUGGGACGAUUUUGAUU .......((((...))))(((((......))))).((((((((.......(((((.(((((......))))))))))......)))))))).............. (-23.12 = -24.08 + 0.96)

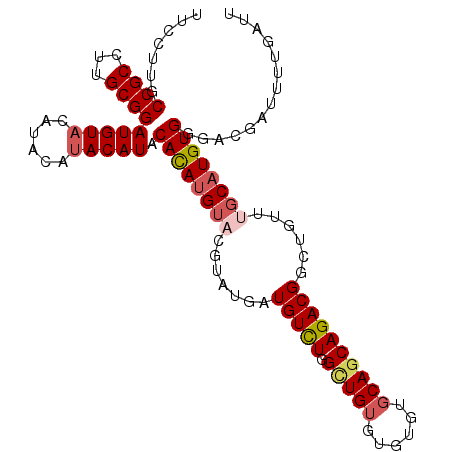

| Location | 19,553,504 – 19,553,599 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 86.88 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -22.10 |
| Energy contribution | -24.10 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19553504 95 - 20766785 AUCUGCAGGCAUACAUAUUUAAGGUGUGAUCGUUUCCUUGCCGCCUUGCGGAUGUUCAUACAUACAUACACAUG--------AUGUUUAGCUGUGUGUGUGCA ((((((((((...(((((.....)))))...((......)).))).))))))).........((((((((((.(--------........))))))))))).. ( -27.20) >DroSec_CAF1 639 103 - 1 AUCUGCAGGCAUACAUAUUUUAGGUGUGAUCGUUUCCUUGCCGCCUUGCGGAUGUACAUACAUACAUACACAUGUACGUAUUAUGUCUGGCUGUGUGUGUGCA ((((((((((...(((((.....)))))...((......)).))).)))))))(((((((((((..((.(((((.......))))).))..))))))))))). ( -33.90) >DroSim_CAF1 512 102 - 1 -UCUGCAGGCAUACAUAUUUAAGGUGUGAUCGUUUCCUUGCCGCCUUGCGGAUGUACAUACAUACAUACACAUGUACGUAUUUUGUCUGGCUGUGUGUGUGCA -...(((.(((((((.......((((.((.....))..))))(((..(((((.((((.(((((........))))).)))))))))..)))))))))).))). ( -32.70) >DroEre_CAF1 690 91 - 1 ------------AUAUAUUUAAGGUGUGAUCGUUUCCUUGCCGCCUUGCGGAUGUACAUACAUACAUACACAUGUACGUAUGAUGUCUGGAUGUGUGUGUGCA ------------.......(((((...........)))))((((...)))).((((((((((((((.(((((((....)))).))).))..)))))))))))) ( -24.70) >DroYak_CAF1 522 102 - 1 AUCCGCAGGCAUACAUAUUUAAGGUGUGAUCGUUUCCUUGCCGCCUUGCGGAUGUACAUACAUACAUACAUAAGUACGUGUGAUGUCUGGUUGUGUGUGU-CA .......((((((((((..(((((((..(........)..).))))))(((((((.(((((.(((........))).))))))))))))..)))))))))-). ( -32.50) >consensus AUCUGCAGGCAUACAUAUUUAAGGUGUGAUCGUUUCCUUGCCGCCUUGCGGAUGUACAUACAUACAUACACAUGUACGUAUGAUGUCUGGCUGUGUGUGUGCA .(((((((((...(((((.....)))))...((......)).))).))))))((((((((((((..((.((((.........)))).))..)))))))))))) (-22.10 = -24.10 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:51 2006