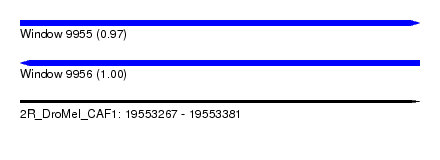
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,553,267 – 19,553,381 |
| Length | 114 |
| Max. P | 0.999656 |

| Location | 19,553,267 – 19,553,381 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 94.80 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -28.56 |
| Energy contribution | -27.92 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19553267 114 + 20766785 UAGAUUUUCCUUUCUUUUUUUCCCGGCGCUCGGAAAGUUCAAGGGUAAGUUCACAAGUGCCGGCAAGUUUAAGGGUGAGAACAAGCCGACAAGAGGGAGAAAGAAAACUAAAAG ..............((((((((((((((((.((((..(........)..))).).))))))(((..((((........))))..))).......)))))))))).......... ( -30.30) >DroSec_CAF1 407 113 + 1 UACAUUUUCCUUCCU-UUUUUCCCGGCGCUCGGAAAGUUCAAGGGUAAGUUCACAAGUGCCGGCAAGUUUAAGGGUGAGAACAAGCCGACAAAAGGGAGAAAGAAAACUAAAAG ....((((.((((((-(((.((((((((((.((((..(........)..))).).))))))))..........(((........))))).))))))))).)))).......... ( -30.90) >DroSim_CAF1 276 113 + 1 UAGAUUUUCCUUCCU-UUUUUCCCGGCGCUCGGAAAGUUCAAGGGUAAGUUCACAAGUGCCGGCAAGUUUAAGGGUGAGAACAAGCCGACAAAAGGGAGAAAGAAAACUAAAAG (((.(((((((((((-(((.((((((((((.((((..(........)..))).).))))))))..........(((........))))).)))))))))...)))))))).... ( -32.60) >DroEre_CAF1 453 113 + 1 UCGAUUUUCCCUUCU-UUUUUCCCGGCACUCGGAAAGUUCAAGGGUAAGUUCACAAGUGCCGGCAAGUUUAAGGGUGAGAACAAGCCGACAAAAGGGAGAAGGAAAACCAAAAG ..(.((((((.....-.(((((((((((((.((((..(........)..))).).))))))(((..((((........))))..))).......))))))))))))).)..... ( -31.90) >DroYak_CAF1 286 113 + 1 CUCAUUUUCCCUUCU-UUUUUCCCGGCAUUCGGAAAGUUCAAGGGUAAGUUCACAAGUGCCGGCAAGUUUAAGGGUGAGAACAAGCCGACAAAAGGGAGAAGGAAAACCAAAAG .((.(((((((((((-(.(((.((((((((.((((..(........)..))).).)))))))).)))...)))(((........))).....))))))))).)).......... ( -29.70) >consensus UAGAUUUUCCUUUCU_UUUUUCCCGGCGCUCGGAAAGUUCAAGGGUAAGUUCACAAGUGCCGGCAAGUUUAAGGGUGAGAACAAGCCGACAAAAGGGAGAAAGAAAACUAAAAG ................((((((((((((((.((((..(........)..))).).))))))(((..((((........))))..))).......))))))))............ (-28.56 = -27.92 + -0.64)

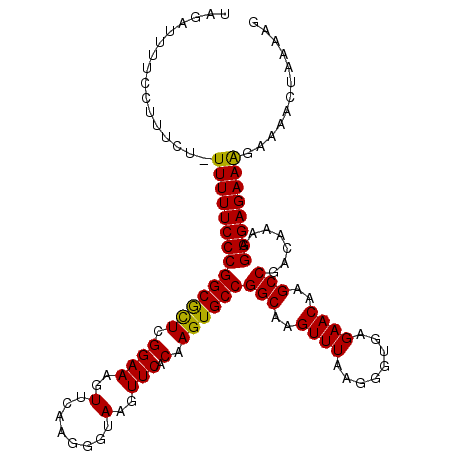
| Location | 19,553,267 – 19,553,381 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 94.80 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -28.74 |
| Energy contribution | -28.22 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.84 |
| SVM RNA-class probability | 0.999656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19553267 114 - 20766785 CUUUUAGUUUUCUUUCUCCCUCUUGUCGGCUUGUUCUCACCCUUAAACUUGCCGGCACUUGUGAACUUACCCUUGAACUUUCCGAGCGCCGGGAAAAAAAGAAAGGAAAAUCUA .......(((((((((((((.......(((..(((..........)))..)))(((.((((.(((..............))))))).)))))))......)))))))))..... ( -28.34) >DroSec_CAF1 407 113 - 1 CUUUUAGUUUUCUUUCUCCCUUUUGUCGGCUUGUUCUCACCCUUAAACUUGCCGGCACUUGUGAACUUACCCUUGAACUUUCCGAGCGCCGGGAAAAA-AGGAAGGAAAAUGUA ......((((((((((((((.......(((..(((..........)))..)))(((.((((.(((..............))))))).)))))))....-..))))))))))... ( -29.94) >DroSim_CAF1 276 113 - 1 CUUUUAGUUUUCUUUCUCCCUUUUGUCGGCUUGUUCUCACCCUUAAACUUGCCGGCACUUGUGAACUUACCCUUGAACUUUCCGAGCGCCGGGAAAAA-AGGAAGGAAAAUCUA ....((((((((((((((((.......(((..(((..........)))..)))(((.((((.(((..............))))))).)))))))....-..))))))))).))) ( -29.54) >DroEre_CAF1 453 113 - 1 CUUUUGGUUUUCCUUCUCCCUUUUGUCGGCUUGUUCUCACCCUUAAACUUGCCGGCACUUGUGAACUUACCCUUGAACUUUCCGAGUGCCGGGAAAAA-AGAAGGGAAAAUCGA ...(((((((((((((((((.......(((..(((..........)))..)))((((((((.(((..............)))))))))))))))....-.))).)))))))))) ( -36.54) >DroYak_CAF1 286 113 - 1 CUUUUGGUUUUCCUUCUCCCUUUUGUCGGCUUGUUCUCACCCUUAAACUUGCCGGCACUUGUGAACUUACCCUUGAACUUUCCGAAUGCCGGGAAAAA-AGAAGGGAAAAUGAG (((....((((((((((......(((((((..(((..........)))..))))))).....................((((((.....))))))...-))))))))))..))) ( -29.80) >consensus CUUUUAGUUUUCUUUCUCCCUUUUGUCGGCUUGUUCUCACCCUUAAACUUGCCGGCACUUGUGAACUUACCCUUGAACUUUCCGAGCGCCGGGAAAAA_AGAAAGGAAAAUCUA .......(((((((((((((.......(((..(((..........)))..)))(((.((((.(((..............))))))).)))))))......)))))))))..... (-28.74 = -28.22 + -0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:48 2006