
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,545,662 – 19,545,791 |
| Length | 129 |
| Max. P | 0.747744 |

| Location | 19,545,662 – 19,545,770 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -28.42 |
| Energy contribution | -28.90 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
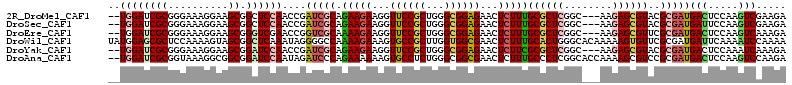
>2R_DroMel_CAF1 19545662 108 + 20766785 --UGGAUCGCGGGAAAGGAAGCGGCUCCAACCGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGC---AAGAGCGUACGCGAUGACUCCAAGUCGAAGA --...((((((.....(....).((((...((((.((((((.((...((((((...))))))...)))))))).)))).---..))))...))))))(((.....)))..... ( -40.30) >DroSec_CAF1 2 108 + 1 --UGGAUCGCGGGAAAGGAAGCGGCUCCAACCGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGC---AAGAGCGUACGCGAUGAUUCCAAGUCGAAGA --(((((((((.....(....).((((...((((.((((((.((...((((((...))))))...)))))))).)))).---..))))...)))))....))))......... ( -39.70) >DroEre_CAF1 3 108 + 1 --UGGAUCGCGGGAAAGGAAGCGGGUCGAACCGGUCGCAAAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGC---AAGAGCGUUCGCGAUGACUCCAAGUCAAAGA --...((((((((........(.(((.(((((..((......)).))))).))).)((((((....))))))((((...---..)))).))))))))(((.....)))..... ( -38.40) >DroWil_CAF1 12300 113 + 1 UAUGGAGCGCUCCAAAAGUAGCGGCUCAAAUAGGGGCCAAAAGAAAGUGCCGCUUGGUGGCGAACUCUUUGCACUGGGCACAAAAAGUGUUCGCGAUGAUUCAAAAUCCAAAA ..((((((........(((.(((((((......))))).(((((...((((((...))))))...))))))))))((((((.....))))))))((....))....))))... ( -36.10) >DroYak_CAF1 2 108 + 1 --UGGAUCGCGGGAAAGGAAGCGGAUCCAACCGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUCGCGCUCGGC---AAGAGCGUACGCGAUGACUCCAAAUCAAAGA --((((((.(((....(((......)))..)))(((((.(((((...((((((...))))))...)))))((((((...---..))))))..))))))).))))......... ( -41.60) >DroAna_CAF1 3 111 + 1 --UGGAUCGCGGUAAAGGCGGCGGAUCCAAUAGAUCCCAGAAAAAAGUGCCUCUGGGCGGCGAACUCUUUGCCCUCGGCACCAAAAGCGUCCGCGAUGACUCCAAGUCCAAGA --((((((((((...(.((.(.(((((.....))))))........(((((...((((((.(....).))))))..))))).....)).)))))))....))))......... ( -38.90) >consensus __UGGAUCGCGGGAAAGGAAGCGGCUCCAACCGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGC___AAGAGCGUACGCGAUGACUCCAAGUCAAAGA ..((((((((..........)).))))))....(((((.(((((...((((((...))))))...)))))((((((........))))))..)))))(((.....)))..... (-28.42 = -28.90 + 0.48)
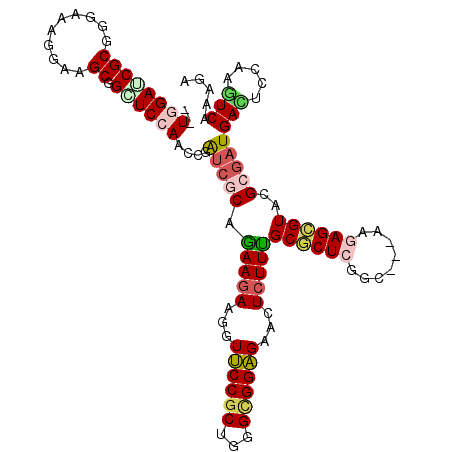

| Location | 19,545,693 – 19,545,791 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.12 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -24.04 |
| Energy contribution | -23.43 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19545693 98 + 20766785 AUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGC---AAGAGCGUACGCGAUGACUCCAAGUCGAAGAUCCUGCCCAUUAAGGGC------AUGU (((((.(((((...((((((...))))))...)))))((((((...---..))))))..)))))(((.....)))........(((((.....))))------)... ( -38.60) >DroSec_CAF1 33 98 + 1 AUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGC---AAGAGCGUACGCGAUGAUUCCAAGUCGAAGAUCCUGCCCAUUAAGGGC------AUGU (((((.(((((...((((((...))))))...)))))((((((...---..))))))..)))))(((.....)))........(((((.....))))------)... ( -36.70) >DroEre_CAF1 34 98 + 1 GUCGCAAAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGC---AAGAGCGUUCGCGAUGACUCCAAGUCAAAGAUCCUGCCCAUUAAGGGC------AUAU (((((....(((.((((.((((((((.(((....))).))))))))---..)))).))))))))(((.....)))........(((((.....))))------)... ( -38.40) >DroWil_CAF1 12333 107 + 1 GGGCCAAAAGAAAGUGCCGCUUGGUGGCGAACUCUUUGCACUGGGCACAAAAAGUGUUCGCGAUGAUUCAAAAUCCAAAAUUUUGCCCAUUAAGACAACAAUAACGG ((((.((((......(((((...)))))(((.((.((((...((((((.....)))))))))).)))))...........))))))))................... ( -26.80) >DroYak_CAF1 33 98 + 1 AUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUCGCGCUCGGC---AAGAGCGUACGCGAUGACUCCAAAUCAAAGAUCCUGCCCAUUAAGGGC------AUGU (((((.(((((...((((((...))))))...)))))((((((...---..))))))..)))))...................(((((.....))))------)... ( -38.80) >DroAna_CAF1 34 101 + 1 AUCCCAGAAAAAAGUGCCUCUGGGCGGCGAACUCUUUGCCCUCGGCACCAAAAGCGUCCGCGAUGACUCCAAGUCCAAGAUACUGCCCAUAAAGUCC------AUAU ....(((......(((((...((((((.(....).))))))..))))).....((....))...(((.....))).......)))............------.... ( -22.40) >consensus AUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGC___AAGAGCGUACGCGAUGACUCCAAGUCAAAGAUCCUGCCCAUUAAGGGC______AUGU (((((.(((((...((((((...))))))...)))))((((((........))))))..)))))(((.....)))......(((........)))............ (-24.04 = -23.43 + -0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:43 2006