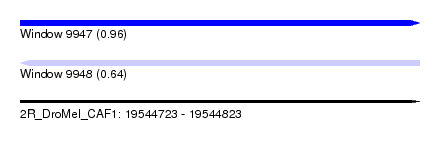
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,544,723 – 19,544,823 |
| Length | 100 |
| Max. P | 0.961665 |

| Location | 19,544,723 – 19,544,823 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.43 |
| Mean single sequence MFE | -35.06 |
| Consensus MFE | -19.01 |
| Energy contribution | -20.32 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
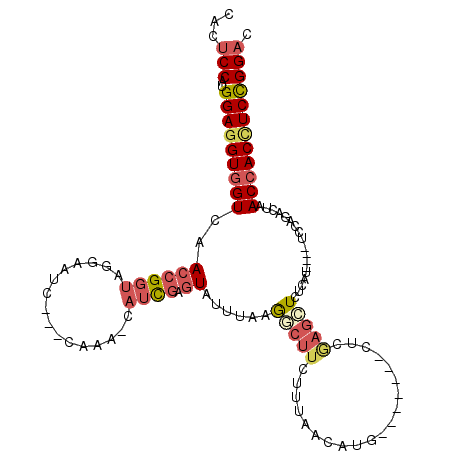
>2R_DroMel_CAF1 19544723 100 + 20766785 GUCCGGAGGUGGUUAGUCUGGA----AUGAGAGCUCGAG-------CAUGUUAAAGAAGCCUUCAAUACUCGAUG-UUUG---GAUUCCUACCGGUUGACCACCUCCAUGGAGUG .((((((((((((((..(.(((----((..(((((((((-------..((((.(((....))).))))))))).)-))).---.)))))....)..)))))))))))..)))... ( -41.90) >DroSec_CAF1 2753 100 + 1 GUCCGGAGGUGGUUAAUCUGGA----AUGAGAGCUCGAG-------CAUUUUAAAGAAGCCUUGAAUACGCGAUG-UUUG---CAUUCCUACCGGUUGACCACCUCCAUGGAGUG .(((((((((((((((((.(((----(((..((((((.(-------.((((..(((....))))))).).))).)-))..---))))))....))))))))))))))..)))... ( -41.50) >DroSim_CAF1 3571 100 + 1 GUCCGGAGGUGGUUAAUCUGGA----AUGAGAGCUCGAG-------CAUGUUAAAGAAGCCUUGAAUACGCGAUG-UUUG---CAUUCCCACCGGUUGACCACCUCCAUGGAGUG .(((((((((((((((((.(((----(((..(((....(-------(.((((.(((....))).)))).))...)-))..---))))))....))))))))))))))..)))... ( -42.20) >DroEre_CAF1 2768 99 + 1 GUCCGGAGGUGGUUAGUCUGGA----AUGAGAACUUAGG-------CACGUUAAAGAAGUCAUAAGUACUCG--GUUUUG---GACUCGUACCGGUUGACCACCUCCAUGGAGUG .(((((((((((((((((..((----(((((.(((((((-------(...........))).))))).))).--))))..---)))(((...))).)))))))))))..)))... ( -38.70) >DroYak_CAF1 3565 101 + 1 GUCCGGAAGUGGUUAGUCUGGA----AUGAGAGCUCAAG-------CACGUUAAAGAAGGCUCAAAUACUCGAUGUUUUG---GAUUCGUACCGGUUUACCACCUCCAUGGAGUG .((((((.(((((....((((.----(((((((((....-------(........)..)))))......((((....)))---)..)))).))))...))))).)))..)))... ( -27.90) >DroPer_CAF1 2194 111 + 1 GACCAGACGUUGUUAAUCUGAGUAGGGGGAAAGGACGAGACAGAGUUAACUUAA----UGUAUAAAUAGUGAAUUUUUUGUGACAAACUUACCUGUUGACCACCUCCAUGGAAUG .((((((.........)))).)).(((((...((....(((((.((........----..((((((..........))))))........)))))))..)).)))))........ ( -18.17) >consensus GUCCGGAGGUGGUUAAUCUGGA____AUGAGAGCUCGAG_______CAUGUUAAAGAAGCCUUAAAUACUCGAUG_UUUG___CAUUCCUACCGGUUGACCACCUCCAUGGAGUG .(((((((((((((((.((((......(((....))).................................(((....)))...........))))))))))))))))..)))... (-19.01 = -20.32 + 1.31)


| Location | 19,544,723 – 19,544,823 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.43 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -12.79 |
| Energy contribution | -13.99 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19544723 100 - 20766785 CACUCCAUGGAGGUGGUCAACCGGUAGGAAUC---CAAA-CAUCGAGUAUUGAAGGCUUCUUUAACAUG-------CUCGAGCUCUCAU----UCCAGACUAACCACCUCCGGAC ...(((..(((((((((.....(((.(((((.---....-..(((((((((((((....)))))..)))-------)))))......))----)))..))).)))))))))))). ( -35.52) >DroSec_CAF1 2753 100 - 1 CACUCCAUGGAGGUGGUCAACCGGUAGGAAUG---CAAA-CAUCGCGUAUUCAAGGCUUCUUUAAAAUG-------CUCGAGCUCUCAU----UCCAGAUUAACCACCUCCGGAC ...(((..(((((((((.((.(....((((((---....-..(((.(((((.(((....)))...))))-------).))).....)))----))).).)).)))))))))))). ( -31.70) >DroSim_CAF1 3571 100 - 1 CACUCCAUGGAGGUGGUCAACCGGUGGGAAUG---CAAA-CAUCGCGUAUUCAAGGCUUCUUUAACAUG-------CUCGAGCUCUCAU----UCCAGAUUAACCACCUCCGGAC ...(((..(((((((((.((.(....((((((---....-..(((.((((..((((...))))...)))-------).))).....)))----))).).)).)))))))))))). ( -31.60) >DroEre_CAF1 2768 99 - 1 CACUCCAUGGAGGUGGUCAACCGGUACGAGUC---CAAAAC--CGAGUACUUAUGACUUCUUUAACGUG-------CCUAAGUUCUCAU----UCCAGACUAACCACCUCCGGAC ...(((..(((((((((.....((((((((((---......--...........)))).......))))-------))..(((.((...----...))))).)))))))))))). ( -27.64) >DroYak_CAF1 3565 101 - 1 CACUCCAUGGAGGUGGUAAACCGGUACGAAUC---CAAAACAUCGAGUAUUUGAGCCUUCUUUAACGUG-------CUUGAGCUCUCAU----UCCAGACUAACCACUUCCGGAC ...(((..(((((((((.....((.......)---)........(((..((..((((.........).)-------))..))..)))..----.........)))))))))))). ( -27.10) >DroPer_CAF1 2194 111 - 1 CAUUCCAUGGAGGUGGUCAACAGGUAAGUUUGUCACAAAAAAUUCACUAUUUAUACA----UUAAGUUAACUCUGUCUCGUCCUUUCCCCCUACUCAGAUUAACAACGUCUGGUC ........((.((.((.(.((((((.(((((........))))).)).(((((....----.))))).....))))...).))...)).))...((((((.......)))))).. ( -14.70) >consensus CACUCCAUGGAGGUGGUCAACCGGUAGGAAUC___CAAA_CAUCGAGUAUUUAAGGCUUCUUUAACAUG_______CUCGAGCUCUCAU____UCCAGACUAACCACCUCCGGAC ...(((..(((((((((..((((((................)))).))......(((((....................)))))..................)))))))))))). (-12.79 = -13.99 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:40 2006