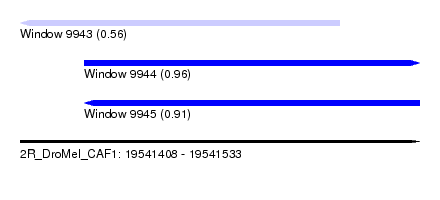
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,541,408 – 19,541,533 |
| Length | 125 |
| Max. P | 0.962041 |

| Location | 19,541,408 – 19,541,508 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.87 |
| Mean single sequence MFE | -23.48 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.86 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19541408 100 - 20766785 UGGAGGCCAAUCAGUUUUCUUAAAUUGUAGAUACAUUUAUAUGUGUUUGUGCUUUAAGAAUUAAAAUAUUGUAUGCCUUUUGCA-AGGUGCGUAUACGGGA------------------- ......((........((((((((.(..((((((((....))))))))..).)))))))).........((((((((((.....-))).))))))).))..------------------- ( -22.40) >DroPse_CAF1 3722 120 - 1 CAGGGGCCAAUCCGUUUUCUUGAAAUGUAGAUACAUUUAUAUGUGUUUGUGGCUUAAGAAUUGAAAUAUUGUAUGCCUUUUGCAAAGGUAAAUAUAGGCAACAUCCAAGUCAAAUUAAAC ...((((((...((((((...))))))(((((((((....))))))))))))))).....((((.....(((.((((((((((....)))))...))))))))......))))....... ( -27.20) >DroSim_CAF1 272 100 - 1 UGGAGGCCAAUCAGUUUUCUUAAAUUGUAGAUACAUUUAUAUGUGUUUGUGCUUUAAGAAUUAAAAUAUUGUAUGCCUUUUGCA-AGGUGCGUAUACGGGA------------------- ......((........((((((((.(..((((((((....))))))))..).)))))))).........((((((((((.....-))).))))))).))..------------------- ( -22.40) >DroYak_CAF1 268 100 - 1 UGGAGGCCAAUCAGUUUUCUUAAAUUGUAGAUACAUUUAUAUGUGUUUGUGCUUUAAGAAUUAAAAUAUUGUAUGCCUUUUGCA-AGGUGCGUAUACGAAA------------------- .((((((....((((.((((((((.(..((((((((....))))))))..).)))))))).......))))...)))))).((.-....))..........------------------- ( -21.40) >DroAna_CAF1 257 101 - 1 UGGAGGCCAAUCAGUUUUCUUAAAUUGUAGAUACAUUUAUAUGUGUUUGUGCUUUAAGAAUUGAAAUAUUGUAUGCCUUUUGCAAAGGUGCGUAUUCGGGA------------------- ......((..(((((..(((((((.(..((((((((....))))))))..).))))))))))))......((((((((((....)))).))))))..))..------------------- ( -24.00) >consensus UGGAGGCCAAUCAGUUUUCUUAAAUUGUAGAUACAUUUAUAUGUGUUUGUGCUUUAAGAAUUAAAAUAUUGUAUGCCUUUUGCA_AGGUGCGUAUACGGGA___________________ .((((((((((.....((((((((.(..((((((((....))))))))..).)))))))).......))))...))))))((((....))))............................ (-19.90 = -19.86 + -0.04)

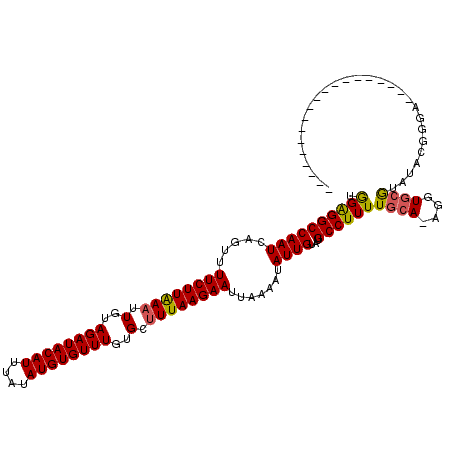
| Location | 19,541,428 – 19,541,533 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.93 |
| Mean single sequence MFE | -19.17 |
| Consensus MFE | -15.19 |
| Energy contribution | -15.83 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.962041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19541428 105 + 20766785 AAAGGCAUACAAUAUUUUAAUUCUUAAAGCACAAACACAUAUAAAUGUAUCUACAAUUUAAGAAAACUGAUUGGCCUCCACCGAUAGUUUUGCCAUCGCUUUCCA--------------- ...((((.............((((((((........((((....))))........))))))))(((((.((((......))))))))).))))...........--------------- ( -18.79) >DroPse_CAF1 3762 119 + 1 AAAGGCAUACAAUAUUUCAAUUCUUAAGCCACAAACACAUAUAAAUGUAUCUACAUUUCAAGAAAACGGAUUGGCCCCUGCCGAUAGUU-UGCCAUCGCUUUCUGUGCGACCCUGUGCGA ...((((.((..................((......((((....)))).(((........)))....))((((((....)))))).)).-)))).((((.......)))).......... ( -22.40) >DroSim_CAF1 292 105 + 1 AAAGGCAUACAAUAUUUUAAUUCUUAAAGCACAAACACAUAUAAAUGUAUCUACAAUUUAAGAAAACUGAUUGGCCUCCACCGAUAGUUUUGCCAUCGCUUUCCA--------------- ...((((.............((((((((........((((....))))........))))))))(((((.((((......))))))))).))))...........--------------- ( -18.79) >DroYak_CAF1 288 105 + 1 AAAGGCAUACAAUAUUUUAAUUCUUAAAGCACAAACACAUAUAAAUGUAUCUACAAUUUAAGAAAACUGAUUGGCCUCCACCGAUAGUUUUGCCAUCGCUUUCCA--------------- ...((((.............((((((((........((((....))))........))))))))(((((.((((......))))))))).))))...........--------------- ( -18.79) >DroAna_CAF1 278 99 + 1 AAAGGCAUACAAUAUUUCAAUUCUUAAAGCACAAACACAUAUAAAUGUAUCUACAAUUUAAGAAAACUGAUUGGCCUCCACCGAUAGUUUUG------CUUUCCA--------------- .((((((.............((((((((........((((....))))........))))))))(((((.((((......))))))))).))------))))...--------------- ( -17.09) >consensus AAAGGCAUACAAUAUUUUAAUUCUUAAAGCACAAACACAUAUAAAUGUAUCUACAAUUUAAGAAAACUGAUUGGCCUCCACCGAUAGUUUUGCCAUCGCUUUCCA_______________ ...((((.............((((((((........((((....))))........))))))))(((((.((((......))))))))).)))).......................... (-15.19 = -15.83 + 0.64)

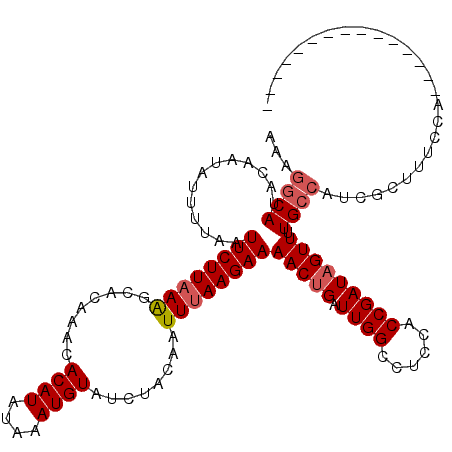
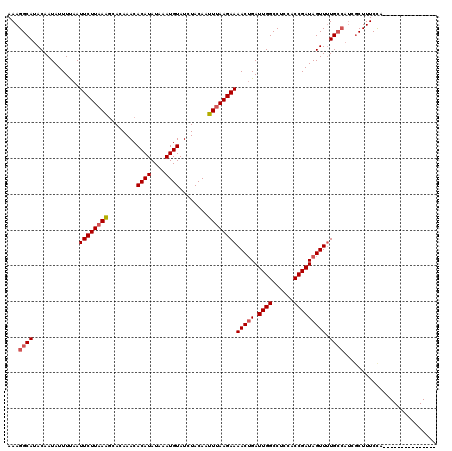
| Location | 19,541,428 – 19,541,533 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.93 |
| Mean single sequence MFE | -26.06 |
| Consensus MFE | -22.08 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
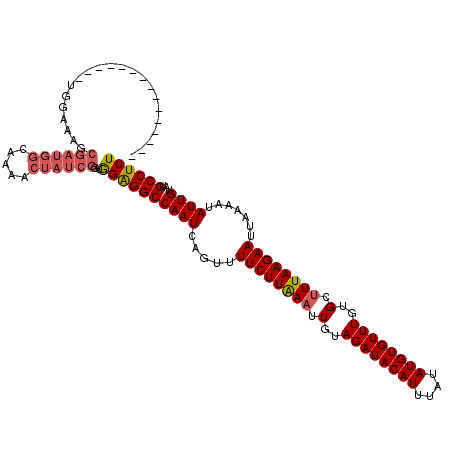
>2R_DroMel_CAF1 19541428 105 - 20766785 ---------------UGGAAAGCGAUGGCAAAACUAUCGGUGGAGGCCAAUCAGUUUUCUUAAAUUGUAGAUACAUUUAUAUGUGUUUGUGCUUUAAGAAUUAAAAUAUUGUAUGCCUUU ---------------.......((((((.....))))))..((((((....((((.((((((((.(..((((((((....))))))))..).)))))))).......))))...)))))) ( -26.00) >DroPse_CAF1 3762 119 - 1 UCGCACAGGGUCGCACAGAAAGCGAUGGCA-AACUAUCGGCAGGGGCCAAUCCGUUUUCUUGAAAUGUAGAUACAUUUAUAUGUGUUUGUGGCUUAAGAAUUGAAAUAUUGUAUGCCUUU .........(((((.......)))))((((-.((....(((....)))........(((((((..(..((((((((....))))))))..)..)))))))..........)).))))... ( -30.10) >DroSim_CAF1 292 105 - 1 ---------------UGGAAAGCGAUGGCAAAACUAUCGGUGGAGGCCAAUCAGUUUUCUUAAAUUGUAGAUACAUUUAUAUGUGUUUGUGCUUUAAGAAUUAAAAUAUUGUAUGCCUUU ---------------.......((((((.....))))))..((((((....((((.((((((((.(..((((((((....))))))))..).)))))))).......))))...)))))) ( -26.00) >DroYak_CAF1 288 105 - 1 ---------------UGGAAAGCGAUGGCAAAACUAUCGGUGGAGGCCAAUCAGUUUUCUUAAAUUGUAGAUACAUUUAUAUGUGUUUGUGCUUUAAGAAUUAAAAUAUUGUAUGCCUUU ---------------.......((((((.....))))))..((((((....((((.((((((((.(..((((((((....))))))))..).)))))))).......))))...)))))) ( -26.00) >DroAna_CAF1 278 99 - 1 ---------------UGGAAAG------CAAAACUAUCGGUGGAGGCCAAUCAGUUUUCUUAAAUUGUAGAUACAUUUAUAUGUGUUUGUGCUUUAAGAAUUGAAAUAUUGUAUGCCUUU ---------------.((...(------(((.......(((....)))..(((((..(((((((.(..((((((((....))))))))..).))))))))))))....))))...))... ( -22.20) >consensus _______________UGGAAAGCGAUGGCAAAACUAUCGGUGGAGGCCAAUCAGUUUUCUUAAAUUGUAGAUACAUUUAUAUGUGUUUGUGCUUUAAGAAUUAAAAUAUUGUAUGCCUUU ......................((((((.....))))))..((((((((((.....((((((((.(..((((((((....))))))))..).)))))))).......))))...)))))) (-22.08 = -23.00 + 0.92)

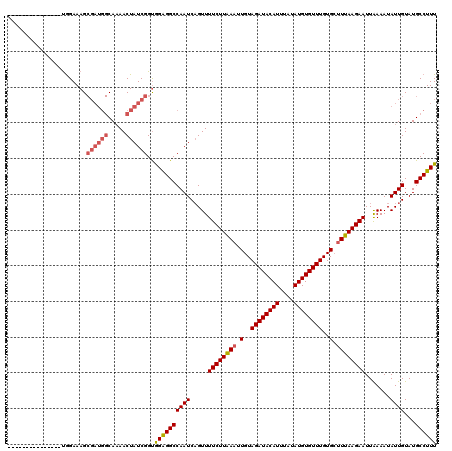
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:36 2006