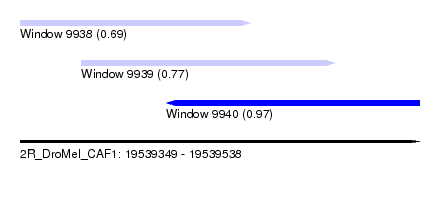
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,539,349 – 19,539,538 |
| Length | 189 |
| Max. P | 0.972918 |

| Location | 19,539,349 – 19,539,458 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 97.43 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -19.89 |
| Energy contribution | -21.75 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
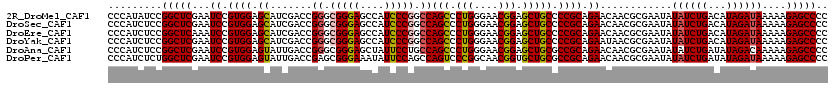
>2R_DroMel_CAF1 19539349 109 + 20766785 GCAAGUAUGAAAAUACAUUUGACAAUAUAGCGAAAUAUUAUGUUUUAUUAAGUUCUAUUGCGAUACAAUGGGGCUCUUUUUAUCUAUGUCAGAUAUAUUCGCGUUGUUC ((..((((....))))((((((((...(((..((((.....))))..)))((((((((((.....))))))))))...........))))))))......))....... ( -24.70) >DroSec_CAF1 4824 109 + 1 GCAAGUAUGAAAAUACAUUUGACAAUAUAGCGAAAUAUUAUGUUUUAUUAAGUUCUAUUGCGAUACAAUGGGGCUCUUUUUAUCUAUGUCAGAUAUAUUCGCGUUGUUC ((..((((....))))((((((((...(((..((((.....))))..)))((((((((((.....))))))))))...........))))))))......))....... ( -24.70) >DroEre_CAF1 4989 109 + 1 GCAAGUAUGAAAAUACAUUUGACAAUAUAGCGAAAUAUUAUGUUUUAUUAAGUUCUAUUGCGAUACAAUGGGGCUCUUUUUAUCUAUGUCAGAUAUAUUCGCGUUGUUC ((..((((....))))((((((((...(((..((((.....))))..)))((((((((((.....))))))))))...........))))))))......))....... ( -24.70) >DroYak_CAF1 4831 109 + 1 GCAAGUAUGAAAAUACAUUUGACAAUAUAGCGAAAUAUUAUGUUUUAUUAAGUUCUAUUGCGAUACAAUGGGGCUCUUUUUAUCUAUGUCAGAUAUAUUCGCGUUAUUC ((..((((....))))((((((((...(((..((((.....))))..)))((((((((((.....))))))))))...........))))))))......))....... ( -24.70) >DroAna_CAF1 4407 109 + 1 GCAAGUAUGAAAAUACAUUUGACAAUAUAGCGAAAUAUUAUGUUUUAUUAAUUUCUAUUGCGAUACAAUGGGGCUCUUUUUGUCUAUAUCAGAUAUAUUCGCGUUGUUC ....((((....))))....((((((...((((((((((...............((((((.....))))))(((.......))).......))))).))))))))))). ( -20.10) >DroPer_CAF1 6938 106 + 1 GCAAGUAUGAAAAUACAUUUGACAAUAUAGCGAAAU---AUGUUUUAUUAACUUCUAUUGCGAUACAAUGGGGCUCUUUUUAUCUAUAUCAGAUAUAUUCGCGUUGUUC ....((((....))))....((((((...(((((..---.............((((((((.....)))))))).......(((((.....)))))..))))))))))). ( -19.60) >consensus GCAAGUAUGAAAAUACAUUUGACAAUAUAGCGAAAUAUUAUGUUUUAUUAAGUUCUAUUGCGAUACAAUGGGGCUCUUUUUAUCUAUGUCAGAUAUAUUCGCGUUGUUC ....((((....))))....((((((...(((((................((((((((((.....)))))))))).....(((((.....)))))..))))))))))). (-19.89 = -21.75 + 1.86)
| Location | 19,539,378 – 19,539,498 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -31.64 |
| Energy contribution | -32.00 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
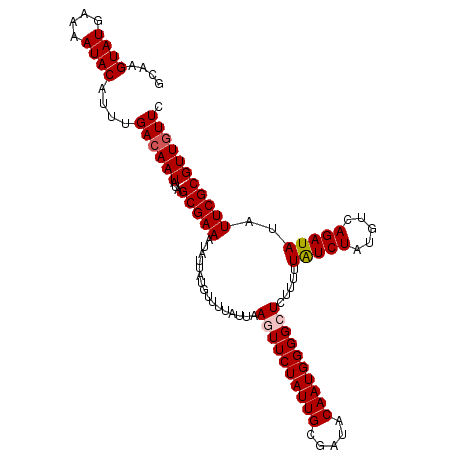
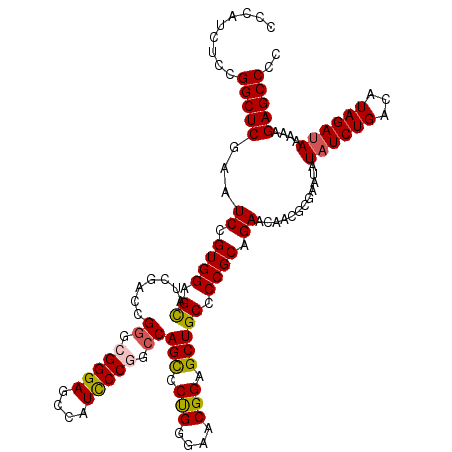
>2R_DroMel_CAF1 19539378 120 + 20766785 GCGAAAUAUUAUGUUUUAUUAAGUUCUAUUGCGAUACAAUGGGGCUCUUUUUAUCUAUGUCAGAUAUAUUCGCGUUGUUCUGCGGGGCAGCUCCGUUCCCAGGGCUGGCCGGGAUGGCUC (((((................((((((((((.....)))))))))).....(((((.....)))))..)))))((..((((...((.((((.((.......)))))).))))))..)).. ( -37.70) >DroSec_CAF1 4853 120 + 1 GCGAAAUAUUAUGUUUUAUUAAGUUCUAUUGCGAUACAAUGGGGCUCUUUUUAUCUAUGUCAGAUAUAUUCGCGUUGUUCUGCGGGGCAGCUCCGUUCCCAGGGCUGGCCGGGAUGGCUC (((((................((((((((((.....)))))))))).....(((((.....)))))..)))))((..((((...((.((((.((.......)))))).))))))..)).. ( -37.70) >DroEre_CAF1 5018 120 + 1 GCGAAAUAUUAUGUUUUAUUAAGUUCUAUUGCGAUACAAUGGGGCUCUUUUUAUCUAUGUCAGAUAUAUUCGCGUUGUUCUGCGGGGCAGCUCCGUUCCCAGGGCUGGCCGGGAUGGCUC (((((................((((((((((.....)))))))))).....(((((.....)))))..)))))((..((((...((.((((.((.......)))))).))))))..)).. ( -37.70) >DroYak_CAF1 4860 120 + 1 GCGAAAUAUUAUGUUUUAUUAAGUUCUAUUGCGAUACAAUGGGGCUCUUUUUAUCUAUGUCAGAUAUAUUCGCGUUAUUCUGCGGGGCAGCUCCGUUCCCAGGGCUGGCCGGGAUGGCUC (((((................((((((((((.....)))))))))).....(((((.....)))))..)))))((((((((...((.((((.((.......)))))).)))))))))).. ( -37.80) >DroAna_CAF1 4436 120 + 1 GCGAAAUAUUAUGUUUUAUUAAUUUCUAUUGCGAUACAAUGGGGCUCUUUUUGUCUAUAUCAGAUAUAUUCGCGUUGUUCUGCGGCGCAGCUCCGUUCCCAGGGCUGGCAGGAAUAGCUC ((((((((((...............((((((.....))))))(((.......))).......))))).)))))((((((((...((.((((.((.......)))))))).)))))))).. ( -34.80) >DroPer_CAF1 6967 117 + 1 GCGAAAU---AUGUUUUAUUAACUUCUAUUGCGAUACAAUGGGGCUCUUUUUAUCUAUAUCAGAUAUAUUCGCGUUGUUCUGCGGCGCAGCACCGUUGCCGGGACUGGCUGGAAUAUUUC (((((..---.............((((((((.....)))))))).......(((((.....)))))..)))))..(((((..((((.(((..(((....)))..)))))))))))).... ( -29.20) >consensus GCGAAAUAUUAUGUUUUAUUAAGUUCUAUUGCGAUACAAUGGGGCUCUUUUUAUCUAUGUCAGAUAUAUUCGCGUUGUUCUGCGGGGCAGCUCCGUUCCCAGGGCUGGCCGGGAUGGCUC (((((................((((((((((.....)))))))))).....(((((.....)))))..)))))((((((((...((.((((((........)))))).)))))))))).. (-31.64 = -32.00 + 0.36)

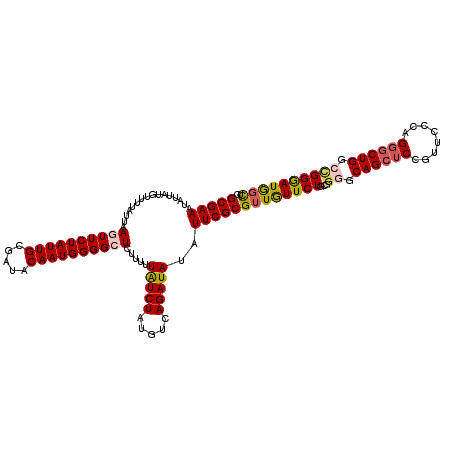

| Location | 19,539,418 – 19,539,538 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -42.65 |
| Consensus MFE | -39.34 |
| Energy contribution | -39.28 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19539418 120 - 20766785 CCCAUAUCCGGCUCGAAUCCGUGGAGCAUCGACCGGGCGGGAGCCAUCCCGGCCAGCCCUGGGAACGGAGCUGCCCCGCAGAACAACGCGAAUAUAUCUGACAUAGAUAAAAAGAGCCCC .........(((((...((.((((.((.......((.(((((....))))).))(((.(((....))).))))).)))).))............((((((...))))))....))))).. ( -43.70) >DroSec_CAF1 4893 120 - 1 CCCAUCUCCGGCUCGAAUCCGUGGAGCAUCGACCGGGCGGGAGCCAUCCCGGCCAGCCCUGGGAACGGAGCUGCCCCGCAGAACAACGCGAAUAUAUCUGACAUAGAUAAAAAGAGCCCC .........(((((...((.((((.((.......((.(((((....))))).))(((.(((....))).))))).)))).))............((((((...))))))....))))).. ( -43.70) >DroEre_CAF1 5058 120 - 1 CCCAUCUCCGGCUCAAAUCCGUGGAGCAUCGACCGGGCGGGAGCCAUCCCGGCCAGCCCUGGGAACGGAGCUGCCCCGCAGAACAACGCGAAUAUAUCUGACAUAGAUAAAAAGAGCCCC .........(((((...((.((((.((.......((.(((((....))))).))(((.(((....))).))))).)))).))............((((((...))))))....))))).. ( -43.70) >DroYak_CAF1 4900 120 - 1 CCCAUCUCCGGCUCGAAUCCGUGGAGCAUCGACCGGGCGGGAGCCAUCCCGGCCAGCCCUGGGAACGGAGCUGCCCCGCAGAAUAACGCGAAUAUAUCUGACAUAGAUAAAAAGAGCCCC .........(((((...((.((((.((.......((.(((((....))))).))(((.(((....))).))))).)))).))............((((((...))))))....))))).. ( -43.70) >DroAna_CAF1 4476 120 - 1 CCCAUCUCCGGCUCGAAUCCGUGGAGUAUUGACCGGGCGGGAGCUAUUCCUGCCAGCCCUGGGAACGGAGCUGCGCCGCAGAACAACGCGAAUAUAUCUGAUAUAGACAAAAAGAGCCCC .........(((((.....((((..((.(((..((((((((((...))))))))(((.(((....))).))).))...))).))..))))......((((...))))......))))).. ( -38.00) >DroPer_CAF1 7004 120 - 1 CCCAUCUCUGGCUCGAAUCCGUGGAGUAUUGACCGAGCGGGAAAUAUUCCAGCCAGUCCCGGCAACGGUGCUGCGCCGCAGAACAACGCGAAUAUAUCUGAUAUAGAUAAAAAGAGCCCC .........(((((.......(((((((((..((....))..)))))))))(((((..(((....)))..))).))(((........)))....((((((...))))))....))))).. ( -43.10) >consensus CCCAUCUCCGGCUCGAAUCCGUGGAGCAUCGACCGGGCGGGAGCCAUCCCGGCCAGCCCUGGGAACGGAGCUGCCCCGCAGAACAACGCGAAUAUAUCUGACAUAGAUAAAAAGAGCCCC .........(((((...((.((((.((.......((.(((((....))))).))(((.(((....))).))))).)))).))............((((((...))))))....))))).. (-39.34 = -39.28 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:31 2006