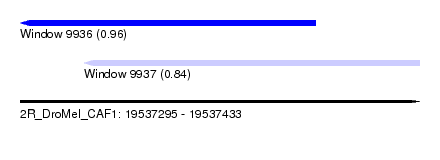
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,537,295 – 19,537,433 |
| Length | 138 |
| Max. P | 0.956789 |

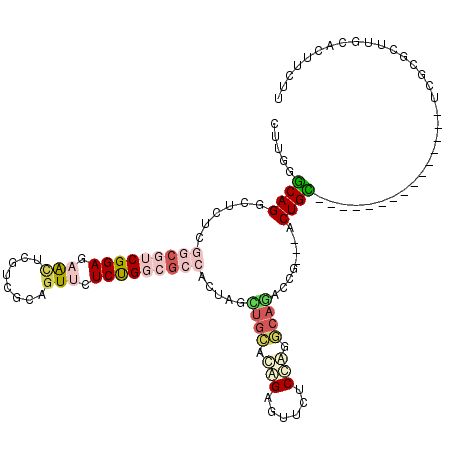
| Location | 19,537,295 – 19,537,397 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.68 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -18.97 |
| Energy contribution | -20.37 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19537295 102 - 20766785 CUUGUGCAGGCUCUCGGCGUCGGAAUACUCGUCGCCGUUCUCUGGCGCCCCUAGCUGCAUGGAGUUGUCCAGGCAGACCG---ACUGC---------------UCGCGCUUGAACUUCUU (..((((..((..(((((((((((..((........))..))))))))).....((((.((((....)))).))))...)---)..))---------------..))))..)........ ( -38.70) >DroPse_CAF1 4429 117 - 1 CUUUGGCAGACUCUCGGCAUCGGAUAGUUCUUCACAGUUUUCCGCCGGGAUUAAAGUUACAGUGCUCUCGGAAUAUUGUU---GCUGCAGCGGGGGGUUAUCGUGACGCUUUGUCUUUUU .......((((....(((.((((((((..(((((((((.(((((..((.(((........))).))..))))).))))).---((....))))))..))))).))).)))..)))).... ( -32.90) >DroSec_CAF1 2773 102 - 1 CUUGUGCAGGCUUUCGGCGUCGGAAUACUCGUCGCCGUUCUCUGGCGCCCCUAGCUGCAUGGAGUUGUCCAUGCAGACCG---ACUGC---------------UCGCGCUUACACUUCUU ...((((..((..(((((((((((..((........))..))))))))).....(((((((((....)))))))))...)---)..))---------------..))))........... ( -42.70) >DroEre_CAF1 2969 102 - 1 CUUGGACAGGCUCUCGGCGUCGGAGAACUCGUCGCAGUUCUCUGGCGCCGCUAGCUGCCCAGAGUUCUCCAGGCAGACCG---ACUGU---------------UCGCGUUUGCACUUUUU .((((...((((..(((((((((((((((......)))))))))))))))..))))..))))..........((((((((---(....---------------))).))))))....... ( -46.50) >DroYak_CAF1 2871 105 - 1 CUUGGACAGGCUCUCGGCGUCGGAGAACUCGUCGCAGUUCUCUGGCGCCACUAGCUGCACGGAGCUCUCCAGGCAAACCGACGACUGU---------------UCGCGUUUGCACUUCUU .(((((..((((((.((((((((((((((......))))))))))))))....(.....))))))).)))))(((((((((.......---------------))).))))))....... ( -45.10) >DroAna_CAF1 2322 117 - 1 CUUUGGCAGGCUCUCCGCAUCGGAGAACUCGUCGCAAUUCUCCGCCUGAAUCAGUUGAACAGCGAUCUCUGCACAAUUUU---GCUGUUGUCCCGCGUUUUCGUCGCGCUUGCACUUCUU .....((((((((((((...))))))...((.(((.((((.......))))......((((((((.............))---)))))).....)))....))....))))))....... ( -30.62) >consensus CUUGGGCAGGCUCUCGGCGUCGGAGAACUCGUCGCAGUUCUCUGGCGCCACUAGCUGCACAGAGUUCUCCAGGCAGACCG___ACUGC_______________UCGCGCUUGCACUUCUU .....((((......(((((((((.(((........))).))))))))).....((((.(((......))).))))........))))................................ (-18.97 = -20.37 + 1.39)

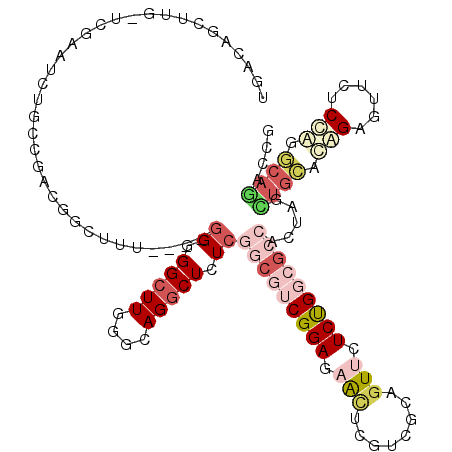
| Location | 19,537,317 – 19,537,433 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.34 |
| Mean single sequence MFE | -46.38 |
| Consensus MFE | -17.05 |
| Energy contribution | -19.25 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19537317 116 - 20766785 UGACAUCUUGAUCGAGUCUGCCGUCGGCUUU----GGGGGCUUGUGCAGGCUCUCGGCGUCGGAAUACUCGUCGCCGUUCUCUGGCGCCCCUAGCUGCAUGGAGUUGUCCAGGCAGACCG ............((.((((((((.(((((..----((((((..((.((((....(((((.(((.....))).)))))...)))))))))))))))))).((((....))))))))))))) ( -51.40) >DroSec_CAF1 2795 116 - 1 UGACAUCUUGAUCGAGUCUGCCGACGGCUUU----GGGGGCUUGUGCAGGCUUUCGGCGUCGGAAUACUCGUCGCCGUUCUCUGGCGCCCCUAGCUGCAUGGAGUUGUCCAUGCAGACCG .........((.(((((...((((((.(...----(..(((((....)))))..)).))))))...)))))))((((.....))))........(((((((((....))))))))).... ( -48.20) >DroEre_CAF1 2991 116 - 1 UGACAGCUUGCUCAGAUCUGCCGACGACUUC----GGGGGCUUGGACAGGCUCUCGGCGUCGGAGAACUCGUCGCAGUUCUCUGGCGCCGCUAGCUGCCCAGAGUUCUCCAGGCAGACCG ..............(.((((((...(((.((----((((((((....)))))))))).)))(((((((((...((((....(((((...)))))))))...))))))))).)))))).). ( -55.50) >DroYak_CAF1 2896 116 - 1 UGACAGCUUGCUUGGAUCUACCGACGACUUC----GGUGGCUUGGACAGGCUCUCGGCGUCGGAGAACUCGUCGCAGUUCUCUGGCGCCACUAGCUGCACGGAGCUCUCCAGGCAAACCG .......(((((((((..((((((.....))----))))((((.(...((((...((((((((((((((......))))))))))))))...))))...).))))..))))))))).... ( -54.90) >DroAna_CAF1 2359 113 - 1 CGACAGUUUG-------CUGUUGUCGGCUUUGUUGGGCGGCUUUGGCAGGCUCUCCGCAUCGGAGAACUCGUCGCAAUUCUCCGCCUGAAUCAGUUGAACAGCGAUCUCUGCACAAUUUU .(.(((.(((-------((((..((((((...(..(((((...((((..(.((((((...)))))).)..)))).......)))))..)...)))))))))))))...))))........ ( -42.10) >DroPer_CAF1 4548 110 - 1 UGAUAACUUU--UGAUUAU-C---CGCUUUU----UGUGGCUUUGGCAGACUCUCGGCAUCGGAUAGUUCUUCACAGUUUUCCGCCGGGAUUAAAGUUACAGUGCUCUCGGAAUAUUGUU ..........--......(-(---((....(----(((((((((((.....(((((((...(((................))))))))))))))))))))))......))))........ ( -26.19) >consensus UGACAGCUUG_UCGAAUCUGCCGACGGCUUU____GGGGGCUUGGGCAGGCUCUCGGCGUCGGAGAACUCGUCGCAGUUCUCUGGCGCCACUAGCUGCACAGAGUUCUCCAGGCAGACCG ...................................((.(((((....))))).))(((((((((.(((........))).))))))))).....((((.(((......))).)))).... (-17.05 = -19.25 + 2.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:28 2006