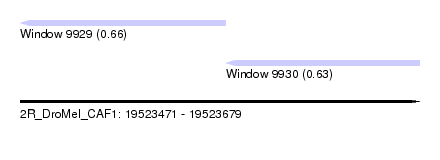
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,523,471 – 19,523,679 |
| Length | 208 |
| Max. P | 0.658407 |

| Location | 19,523,471 – 19,523,578 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.07 |
| Mean single sequence MFE | -24.14 |
| Consensus MFE | -12.79 |
| Energy contribution | -12.83 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
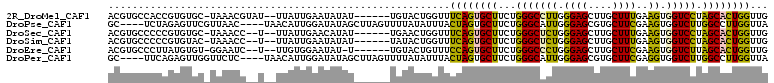
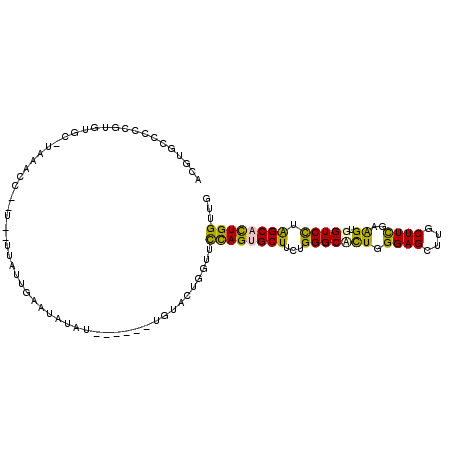
>2R_DroMel_CAF1 19523471 107 - 20766785 UGUAUCCCUAUUACGGAUGGCGAGGUCUCUUGGCCCUUUCGGCUACACCCCUACUCAUUUUUACCAUCCUAUCGCCGGUUCGUAUUCCU--CGCUCUCUUCAGA-------AAUA--U .(((.......)))(((.(((((((.....(((((.....)))))................(((...((.......))...)))..)))--)))).))).....-------....--. ( -21.50) >DroSec_CAF1 547 107 - 1 UCUACCCCUAUUACGGAUGGCGGGGUCUCUUGGCUCUUUCGGCUACACCCCUUCUCAUUUUUACCAUCCUAUCACCGGUAGGUAUCCCU--UGCUCCCUUCCGA-------AAUC--U .............((((.((((((((....(((((.....))))).)))))................(((((.....))))).......--.)))....)))).-------....--. ( -25.20) >DroSim_CAF1 553 107 - 1 UCUAUCCCUAUUACGGAUGGCGGGGUCUCUUGGCUCUUUCGGCUACACCCCUACUGAUUUUUACCAUCCUAUCACCGGUAGGAAUCCCU--UGCUCUCUUCCGA-------AAUC--U .............((((.((((((((....(((((.....))))).)))))....((((((((((...........))))))))))...--.)))....)))).-------....--. ( -29.60) >DroEre_CAF1 565 112 - 1 UCUACCCCUAUUAUGGAUGGCGUGGUCUCCUGGCUCUUUCGGCUACACCCCUACUAAUCUUCACCAUCCUAUCGCCGGUGGGUAUAGCU--UGUG--CUUAAGAUCAAUCAAAUC--G ...............(((....((((((...(((.(....(((((.((((............(((...........))))))).)))))--.).)--))..)))))).....)))--. ( -22.90) >DroYak_CAF1 563 107 - 1 UCUACCCCUAUUACGGAUGGCGUGGUCUCUUGGCACUUUCGGCUACACCCCUACUGAUCUUCACCAUCCUAUCACCGGUAGAUAUACCU--------CUUAAGG---AACAUAUCGUG ((((((.....((.((((((.(.((((....((.......((.....))))....))))..).)))))))).....))))))....(((--------....)))---........... ( -23.41) >DroAna_CAF1 597 109 - 1 UGUAUCCAUAUUAUGGUUGGCGUGGACUUCUGGGACUCUCAGCCACGCCCUUAUUGAUAUUCACUCUUCUUUCCCCGGUAAUAAUAUUUAUAAACAAGAAUAGA-------AAUA--U .......((((((.....(((((((....(((((...)))))))))))).....))))))...((.(((((......((((......))))....))))).)).-------....--. ( -22.20) >consensus UCUACCCCUAUUACGGAUGGCGUGGUCUCUUGGCUCUUUCGGCUACACCCCUACUGAUUUUCACCAUCCUAUCACCGGUAGGUAUACCU__UGCUC_CUUCAGA_______AAUC__U ((((((........((((((.((((.....(((((.....))))).....)))).........)))))).......)))))).................................... (-12.79 = -12.83 + 0.03)

| Location | 19,523,578 – 19,523,679 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 71.27 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -19.91 |
| Energy contribution | -18.67 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.47 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19523578 101 - 20766785 ACGUGCCACCGUGUGC-UAAACGUAU--UUAUUGAAUAUAU------UGUACUGGUUUCAGUGCUUCUGGGCCUUGGGAGCUUGCUUUGAAGUGGUCCUAGCACUGGUUG .......((((.((((-.....((((--((...))))))..------.)))))))).((((((((...(((((((.((((....)))).))..))))).))))))))... ( -30.40) >DroPse_CAF1 659 102 - 1 GC----UCUAGAGUUCGUUAAC----UAACAUUGGAUAUAGCUUAGUUUUAUAUUUACUAGUGCUUCUGGGCAUUGGGAGCGUGCUUCGAAGUGGUCUUGGCCUUGGUUA ((----(.(..(((.((((...----......(((((((((.......)))))))))((((((((....)))))))).)))).)))..).)))(((....)))....... ( -23.40) >DroSec_CAF1 654 99 - 1 ACGUGCCCCCGUGUGC-UAAACC--U--UUAUUGAACAUAU------UGAACUGGUUUCAGUGCUUCUGGGCUCUGGGAGCUUGCUUUGAAGUGGUCCUAGCACUGGUUG ........(((.((((-((.(((--.--.............------...((((....))))(((((..(((...(....)..)))..))))))))..)))))))))... ( -26.40) >DroSim_CAF1 660 99 - 1 ACGUGCCCCCGUGUAC-UAAACC--U--UUAUUGAAUAUAU------UAUACUGGUUUCAGUGCUUCUGGGCUCUGGGAGCUUGCUUUGAAGUGGUCCUAGCACUGGUUG ..((((......))))-.(((((--.--.(((.........------.)))..)))))(((((((...(((((((.((((....))))..)).))))).))))))).... ( -26.50) >DroEre_CAF1 677 98 - 1 ACGUGCCCUUAUGUGU-GGAAUC--U--UUGUGGAAUAU-U------UGUACUGUUUCCAGUGCUUCUGGGCCCUGGGAGCUUGCUUCGAAGUGGUCUUAGCACUGGUUG ..((((((........-))..((--(--....)))....-.------.)))).....((((((((...(((((((.((((....))))..)).))))).))))))))... ( -31.20) >DroPer_CAF1 569 102 - 1 GC----UUCAGAGUUGGUUCUC----UAACAUUGGAUAUAGCUUAGUUUUAUAUUUACUAGUGCUUCUGGGCAUUGGGAGCGUGCUUCGAGGUGGUCUUGGCCUUGGUUA ((----(((..(((..((((..----......(((((((((.......)))))))))((((((((....))))))))))))..)))..)))))(((....)))....... ( -28.30) >consensus ACGUGCCCCCGUGUGC_UAAACC__U__UUAUUGAAUAUAU______UGUACUGGUUCCAGUGCUUCUGGGCACUGGGAGCUUGCUUCGAAGUGGUCCUAGCACUGGUUG .........................................................((((((((...(((((((.((((....))))..)).))))).))))))))... (-19.91 = -18.67 + -1.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:21 2006