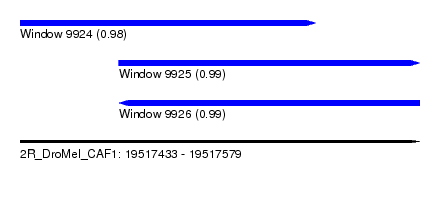
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,517,433 – 19,517,579 |
| Length | 146 |
| Max. P | 0.993516 |

| Location | 19,517,433 – 19,517,541 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -47.05 |
| Consensus MFE | -40.79 |
| Energy contribution | -41.10 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19517433 108 + 20766785 GCAACCUCUUUUACUGGUGAAGUAACCACUUGCGGUUGUGGUGGAGGAGUCGCUACCUUCGCAGGAGUAAUCUCCUUCGGAGGUGGUGGGGAAGACACUACCUUCACU ((((((.(......((((......))))...).))))))((((((((((((((((((((((.(((((....))))).)))))))))))(......)))).)))))))) ( -48.00) >DroSec_CAF1 14043 108 + 1 GCAACCUCUUUUACUGGUGAAGUAACCACUUGCGGUUGUGGUGGAGGAGUCUUUACCUUCGCAGGAGUAAUCUCCUUCGGAGGUGGUGGGGAAGACACGACCUUCACU ((((((.(......((((......))))...).))))))((((((((......((((((((.(((((....))))).))))))))(((.......)))..)))))))) ( -43.70) >DroSim_CAF1 14816 108 + 1 GCAACCUCUUUUACUGGUGAAGUAACCACUGGCGGUUGUGGUGGAGGAGUCACUACCUUCGCAGGAGUAAUCUCCUUCGGAGGUGGUGGGGAAGACACUACCUUCACU ((((((.((.....((((......))))..)).))))))((((((((((((((((((((((.(((((....))))).)))))))))))(......)))).)))))))) ( -50.00) >DroYak_CAF1 14126 108 + 1 GCAACCUCCUUUACUGGAGAAGUAACCACUUGCGGCGGUGGUGGAGGAGUCACUACCUUCGCAGGAGUAACCUCCUUCGGUGGCGGUGGAGGUGUCACUACCUUCACU .....((((......))))......((((((((((.((((((((.....)))))))).)))))((((....))))...))))).((((((((((....)))))))))) ( -46.50) >consensus GCAACCUCUUUUACUGGUGAAGUAACCACUUGCGGUUGUGGUGGAGGAGUCACUACCUUCGCAGGAGUAAUCUCCUUCGGAGGUGGUGGGGAAGACACUACCUUCACU ....(((((..((((.....))))(((((........))))))))))..((((((((((((.(((((....))))).)))))))))))).((((.......))))... (-40.79 = -41.10 + 0.31)


| Location | 19,517,469 – 19,517,579 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 92.73 |
| Mean single sequence MFE | -50.05 |
| Consensus MFE | -45.79 |
| Energy contribution | -46.48 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.93 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19517469 110 + 20766785 UGUGGUGGAGGAGUCGCUACCUUCGCAGGAGUAAUCUCCUUCGGAGGUGGUGGGGAAGACACUACCUUCACUGGAGUAACCUCCUUGACUUGAGCUUGUGGUGGAGGAGG ...((((((((((((((((((((((.(((((....))))).)))))))))))(......)))).)))))))).......(((((((.(((.........))).))))))) ( -51.90) >DroSec_CAF1 14079 110 + 1 UGUGGUGGAGGAGUCUUUACCUUCGCAGGAGUAAUCUCCUUCGGAGGUGGUGGGGAAGACACGACCUUCACUGGAGUAACCUCCUUCACUUGAGCUUGUGGCGGAGGAGG ...((((((((......((((((((.(((((....))))).))))))))(((.......)))..)))))))).......(((((.((((........)))).)))))... ( -47.30) >DroSim_CAF1 14852 110 + 1 UGUGGUGGAGGAGUCACUACCUUCGCAGGAGUAAUCUCCUUCGGAGGUGGUGGGGAAGACACUACCUUCACUGGAGUAACCUCCUUCACUUGAGAUUGUGGCGGAGGAGG ...((((((((((((((((((((((.(((((....))))).)))))))))))(......)))).)))))))).......(((((.((((........)))).)))))... ( -52.00) >DroYak_CAF1 14162 110 + 1 GGUGGUGGAGGAGUCACUACCUUCGCAGGAGUAACCUCCUUCGGUGGCGGUGGAGGUGUCACUACCUUCACUGGAGUAACCUCCUUCACUUGAGCUUGUGGUGGAGGAGA ((((((((.....)))))))).(((.(((((....))))).)))...(((((((((((....))))))))))).......((((((((((.........)))))))))). ( -49.00) >consensus UGUGGUGGAGGAGUCACUACCUUCGCAGGAGUAAUCUCCUUCGGAGGUGGUGGGGAAGACACUACCUUCACUGGAGUAACCUCCUUCACUUGAGCUUGUGGCGGAGGAGG ...((((((((...(((((((((((.(((((....))))).)))))))))))(......)....)))))))).......(((((.((((........)))).)))))... (-45.79 = -46.48 + 0.69)

| Location | 19,517,469 – 19,517,579 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 92.73 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -31.35 |
| Energy contribution | -31.72 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19517469 110 - 20766785 CCUCCUCCACCACAAGCUCAAGUCAAGGAGGUUACUCCAGUGAAGGUAGUGUCUUCCCCACCACCUCCGAAGGAGAUUACUCCUGCGAAGGUAGCGACUCCUCCACCACA ..........................(((((...((((..((.((((.(((.......))).)))).))..)))).....((((((....)))).))..)))))...... ( -30.00) >DroSec_CAF1 14079 110 - 1 CCUCCUCCGCCACAAGCUCAAGUGAAGGAGGUUACUCCAGUGAAGGUCGUGUCUUCCCCACCACCUCCGAAGGAGAUUACUCCUGCGAAGGUAAAGACUCCUCCACCACA ...(((((..(((........)))..)))))........(((..((..(.(((((.......((((.((.(((((....))))).)).)))).))))).)..))..))). ( -34.20) >DroSim_CAF1 14852 110 - 1 CCUCCUCCGCCACAAUCUCAAGUGAAGGAGGUUACUCCAGUGAAGGUAGUGUCUUCCCCACCACCUCCGAAGGAGAUUACUCCUGCGAAGGUAGUGACUCCUCCACCACA ...(((((..(((........)))..)))))........(((.((((.(((.......))).))))..(.(((((.((((((((....))).)))))))))).)..))). ( -35.70) >DroYak_CAF1 14162 110 - 1 UCUCCUCCACCACAAGCUCAAGUGAAGGAGGUUACUCCAGUGAAGGUAGUGACACCUCCACCGCCACCGAAGGAGGUUACUCCUGCGAAGGUAGUGACUCCUCCACCACC .....................(((.(((((((((((((......)).)))))).....(((..((..((.(((((....))))).))..))..))).))))).))).... ( -37.60) >consensus CCUCCUCCACCACAAGCUCAAGUGAAGGAGGUUACUCCAGUGAAGGUAGUGUCUUCCCCACCACCUCCGAAGGAGAUUACUCCUGCGAAGGUAGUGACUCCUCCACCACA ((((((.(((...........))).))))))........(((..(((.(((.......))).)))...(.(((((.((((((((....))).)))))))))).)..))). (-31.35 = -31.72 + 0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:17 2006