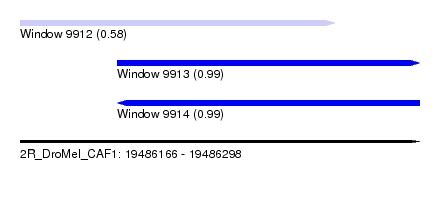
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,486,166 – 19,486,298 |
| Length | 132 |
| Max. P | 0.989235 |

| Location | 19,486,166 – 19,486,270 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.91 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -22.75 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19486166 104 + 20766785 GAUUGAAAUAACUC--------UUUUAUGGAAAAUAAGGAAGGCGUAAGGCACUUUUAUGCCAAUGCAACUGAACUAACCGCAAGUCCGCUCCUGACUCGUCGAGUCAGGAA-------- ............((--------(.....)))......(((.((((((((.....))))))))..(((.............)))..)))..(((((((((...))))))))).-------- ( -26.72) >DroSec_CAF1 2742 104 + 1 AAUUGUAAUAACUC--------UUUUAUGGAAAAUAGGGAAGGCAUAAGUCACUUUUAUGCCAAUGCAACUGAACUAACCGCAAGUCCGCUCCUGACUCGUCGAGUCAGGAA-------- ..(((((....(((--------(.((......)).))))..((((((((.....))))))))..)))))...........((......))(((((((((...))))))))).-------- ( -29.40) >DroSim_CAF1 3026 104 + 1 AAUUGUAAUAACUC--------UUUUAUGGAAAAUAAGGAAGGCAUAAGUCACUUUUAUGCCAAUGCAACUGAACUAACCGCAAGUCCGCUCCUGACUCGUCGAGUCAGGAA-------- ..(..(((......--------..)))..).......(((.((((((((.....))))))))..(((.............)))..)))..(((((((((...))))))))).-------- ( -28.92) >DroEre_CAF1 2925 118 + 1 GAUUGUAAUAACUUGAAAAUUGUUAAAUGAAAACCAAGGAAGGCAUGAGCUGCUCUUGUACCGCUGC--CCGCACUAACCGCAAGUCCGCUCCUGACCCAUCGAGUCUGGCCGCACACAC ...(((....(((((......((((.........(((((..(((....)))..)))))....((...--..))..))))..)))))..((.((.(((.......))).))..)))))... ( -18.90) >consensus AAUUGUAAUAACUC________UUUUAUGGAAAAUAAGGAAGGCAUAAGUCACUUUUAUGCCAAUGCAACUGAACUAACCGCAAGUCCGCUCCUGACUCGUCGAGUCAGGAA________ .....................................(((.((((((((.....)))))))).................(....))))..(((((((((...)))))))))......... (-22.75 = -23.00 + 0.25)


| Location | 19,486,198 – 19,486,298 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 95.71 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -30.39 |
| Energy contribution | -30.50 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19486198 100 + 20766785 AGGCGUAAGGCACUUUUAUGCCAAUGCAACUGAACUAACCGCAAGUCCGCUCCUGACUCGUCGAGUCAGGAA-ACUCUCGCAAAAACCUGCGCUCUUUCGG .((((((((.....))))))))................(((.(((...(((((((((((...))))))))).-......(((......))))).))).))) ( -31.30) >DroSec_CAF1 2774 101 + 1 AGGCAUAAGUCACUUUUAUGCCAAUGCAACUGAACUAACCGCAAGUCCGCUCCUGACUCGUCGAGUCAGGAACACUCUCGCAGAAACCUGCGCUCUUUCUC .((((((((.....))))))))...((....((......(....)...(.(((((((((...))))))))).)....))((((....))))))........ ( -32.20) >DroSim_CAF1 3058 101 + 1 AGGCAUAAGUCACUUUUAUGCCAAUGCAACUGAACUAACCGCAAGUCCGCUCCUGACUCGUCGAGUCAGGAACACUCUCGCAGAAACCUGCGCUCUUUCCC .((((((((.....))))))))...((....((......(....)...(.(((((((((...))))))))).)....))((((....))))))........ ( -32.20) >consensus AGGCAUAAGUCACUUUUAUGCCAAUGCAACUGAACUAACCGCAAGUCCGCUCCUGACUCGUCGAGUCAGGAACACUCUCGCAGAAACCUGCGCUCUUUCCC .((((((((.....))))))))...((....((.......((......))(((((((((...)))))))))......))((((....))))))........ (-30.39 = -30.50 + 0.11)

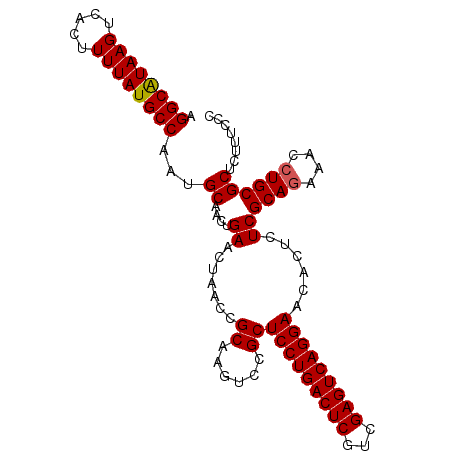
| Location | 19,486,198 – 19,486,298 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 95.71 |
| Mean single sequence MFE | -40.40 |
| Consensus MFE | -38.23 |
| Energy contribution | -38.90 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19486198 100 - 20766785 CCGAAAGAGCGCAGGUUUUUGCGAGAGU-UUCCUGACUCGACGAGUCAGGAGCGGACUUGCGGUUAGUUCAGUUGCAUUGGCAUAAAAGUGCCUUACGCCU ..(((.((.(((((((((..((....))-((((((((((...)))))))))).))))))))).))..)))....((...(((((....)))))....)).. ( -36.70) >DroSec_CAF1 2774 101 - 1 GAGAAAGAGCGCAGGUUUCUGCGAGAGUGUUCCUGACUCGACGAGUCAGGAGCGGACUUGCGGUUAGUUCAGUUGCAUUGGCAUAAAAGUGACUUAUGCCU ..(((.((.(((((((((..((....))(((((((((((...)))))))))))))))))))).))..))).........(((((((.......))))))). ( -41.80) >DroSim_CAF1 3058 101 - 1 GGGAAAGAGCGCAGGUUUCUGCGAGAGUGUUCCUGACUCGACGAGUCAGGAGCGGACUUGCGGUUAGUUCAGUUGCAUUGGCAUAAAAGUGACUUAUGCCU .(((..((.(((((((((..((....))(((((((((((...)))))))))))))))))))).))..))).........(((((((.......))))))). ( -42.70) >consensus GAGAAAGAGCGCAGGUUUCUGCGAGAGUGUUCCUGACUCGACGAGUCAGGAGCGGACUUGCGGUUAGUUCAGUUGCAUUGGCAUAAAAGUGACUUAUGCCU ..(((.((.(((((((((..((....))(((((((((((...)))))))))))))))))))).))..))).........(((((((.......))))))). (-38.23 = -38.90 + 0.67)

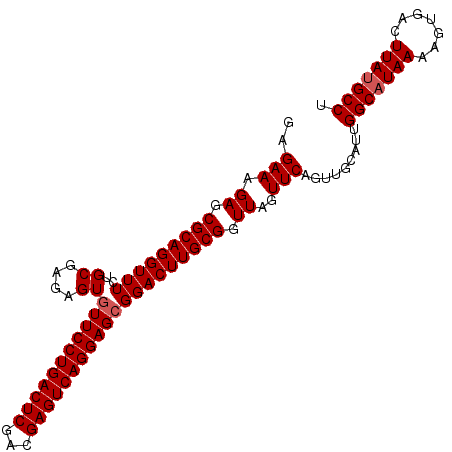
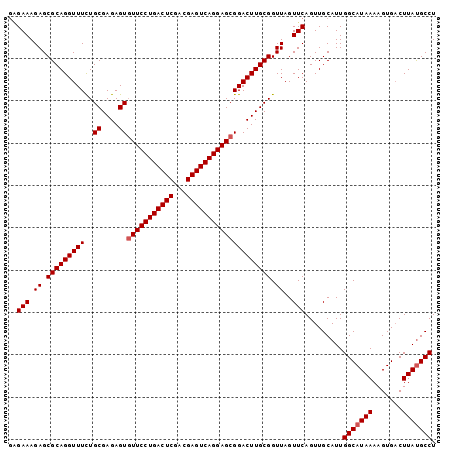
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:05 2006