
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,472,493 – 19,472,653 |
| Length | 160 |
| Max. P | 0.813584 |

| Location | 19,472,493 – 19,472,613 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -45.80 |
| Consensus MFE | -38.67 |
| Energy contribution | -38.23 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
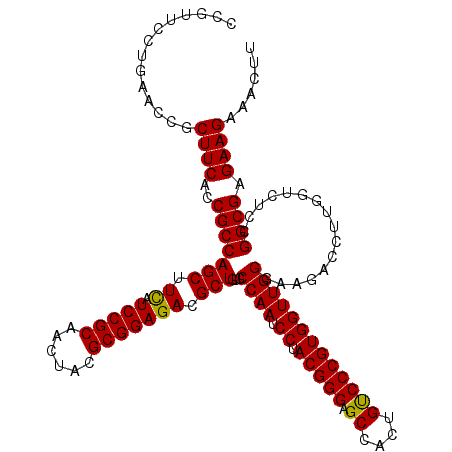
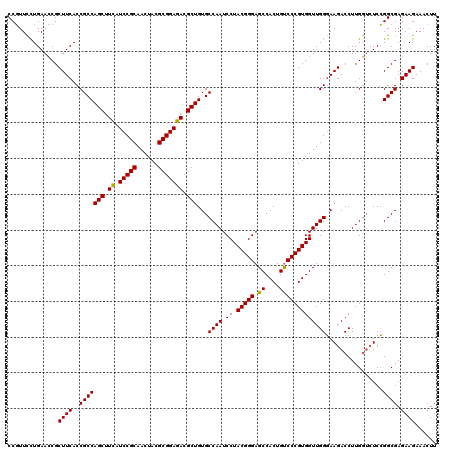
>2R_DroMel_CAF1 19472493 120 + 20766785 CCGUUCUUGAACCGCUUCACCGCCAGCUUCAUCCGCAACUACGCGGAGACGCUGUGCCAAUCCUACGGGAGCCACUGUCCCGUGGUUGGGAAGACCUUGGUAUCCGGCGAGAAGAAACUU ...((((((..(((((((.((((((((.((.(((((......))))))).)))).)).....((((((((.......))))))))..))))))((....))...)))))))))....... ( -45.00) >DroSim_CAF1 2316 120 + 1 CCGUUCCUGAACCGCUUCACCGCCAGCUUCAUCCGCAACUACGCGGAGACGCUGUGCCAAUCCUACGGGAGCCACUGUCCCGUGGUUGGGAAGACCUUGGUCUCCGGCGAGAAGAAACUU ..............((((.....((((.((.(((((......))))))).))))((((....((((((((.......))))))))...(((.(((....)))))))))).))))...... ( -45.20) >DroYak_CAF1 1623 120 + 1 CCCUUUCUGAAUCGCUUCACCGCCAGCUUUAUCCGCAACUACGCGGAGACGCUGUGCCAAUCCUACGGGAGCCACUGCCCCGUGGUUGGCAAGACCUUGAGCUCGGGCGAGAAGAAACUU ...(((((...((((((....((((((....(((((......)))))...))))((((((.((.(((((.((....))))))))))))))).........))..))))))..)))))... ( -47.20) >consensus CCGUUCCUGAACCGCUUCACCGCCAGCUUCAUCCGCAACUACGCGGAGACGCUGUGCCAAUCCUACGGGAGCCACUGUCCCGUGGUUGGGAAGACCUUGGUCUCCGGCGAGAAGAAACUU ..............((((..(((((((.((.(((((......))))))).)))...((((.((.(((((.((....)))))))))))))................)))).))))...... (-38.67 = -38.23 + -0.44)

| Location | 19,472,533 – 19,472,653 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -50.23 |
| Consensus MFE | -44.71 |
| Energy contribution | -44.27 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
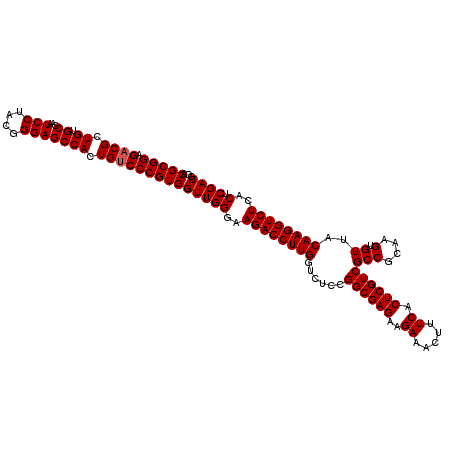
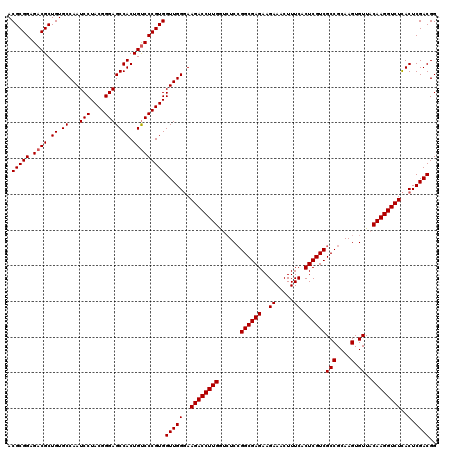
>2R_DroMel_CAF1 19472533 120 + 20766785 ACGCGGAGACGCUGUGCCAAUCCUACGGGAGCCACUGUCCCGUGGUUGGGAAGACCUUGGUAUCCGGCGAGAAGAAACUUUCACUCGUCGCCGCAAGUGUUACAAGGUCUCACUCGACGG .(((((.((((.((.((...(((....))))))).)))))))))((((((.((((((((.....(((((((..((.....)).))))))).(....).....))))))))..)))))).. ( -49.80) >DroSim_CAF1 2356 120 + 1 ACGCGGAGACGCUGUGCCAAUCCUACGGGAGCCACUGUCCCGUGGUUGGGAAGACCUUGGUCUCCGGCGAGAAGAAACUUUCACUCGUCGCCGCAAGUGUUACAAGGUCUCACUCGACGG .(((((.((((.((.((...(((....))))))).)))))))))((((((.((((((((.....(((((((..((.....)).))))))).(....).....))))))))..)))))).. ( -49.80) >DroYak_CAF1 1663 120 + 1 ACGCGGAGACGCUGUGCCAAUCCUACGGGAGCCACUGCCCCGUGGUUGGCAAGACCUUGAGCUCGGGCGAGAAGAAACUUUCACUCGUCGCCGCAAGUGUUACAAGGUCUUACUCGACGG ..(((....)))...(((((.((.(((((.((....))))))))))))))(((((((((.(((.((((((..((.........))..))))).).)))....)))))))))......... ( -51.10) >consensus ACGCGGAGACGCUGUGCCAAUCCUACGGGAGCCACUGUCCCGUGGUUGGGAAGACCUUGGUCUCCGGCGAGAAGAAACUUUCACUCGUCGCCGCAAGUGUUACAAGGUCUCACUCGACGG ..(((....))).(((((((.((.(((((.((....)))))))))))))..((((((((......((((((..((.....)).))))))(((....).))..)))))))))))....... (-44.71 = -44.27 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:58 2006