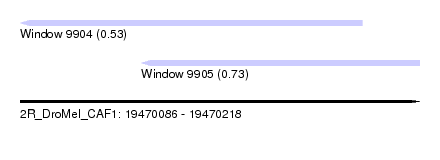
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,470,086 – 19,470,218 |
| Length | 132 |
| Max. P | 0.725905 |

| Location | 19,470,086 – 19,470,199 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.17 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -15.32 |
| Energy contribution | -17.77 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.47 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19470086 113 - 20766785 CUAUUAUGGACUAUGGG---CCAUG-AGGCCAACUUCUUU---GAGUUCGUCUUUUCCCACGCUAAACUGCGAAAUUGAAUGUGAGAAAAUUUGCAUGGGUCAGGCGGAUUCGGAACGUA ....((((.......((---((...-.))))......(((---((((((((((...(((.(((......))).......(((..((....))..))))))..))))))))))))).)))) ( -35.70) >DroSec_CAF1 4768 113 - 1 CUAUUAUGGACUAUGGG---CCAUG-GGCCCAACUUCUUU---GAGUUCGUCUUUUCGCACGCUAAACUGCGAAACCGAAUGUGAGAAAAUUUGCAUGGGUCAGGCGGAUUCGGAACGUA ....((((.....((((---((...-)))))).....(((---((((((((((.((((((........))))))(((..(((..((....))..))).))).))))))))))))).)))) ( -37.90) >DroSim_CAF1 6015 113 - 1 CUAUUAUGGGCUAUGGG---CCAUG-AGGCCAACUUCUUU---GAGUUCGUCUUUUCCCACGCUAAACUGCGAAACCGAAUGUGAGAAAAUUUGCAUGGGUCAGGCGGAUUCGGAACGUA ....((((..((...((---((...-.)))).........---((((((((((...(((.(((......))).......(((..((....))..))))))..))))))))))))..)))) ( -36.50) >DroPer_CAF1 7083 97 - 1 -----------UAUAGUCAACAAUGCUAACCAACUUCUUACUCGCAUGUUACUUGUCCCACACUCAAUGG---ACAUAAAUGUGGGAAAAGUAACACGGGUUC---------CACUUGCA -----------..((((.......))))..(((......(((((..((((((((.(((((((........---.......))))))).)))))))))))))..---------...))).. ( -23.96) >DroEre_CAF1 4692 111 - 1 CAAUUAUUGGCA--GAA---CCAUG-AGGCCAACUUCUUU---GAGUUCGUCUUUUCCCACGCUAAACUGCGAAAUCAAAUGUGAGAAAAUUUGCAUGGGUCAGGCGGAUUCGGAACGUA ......(((((.--...---.....-..)))))....(((---((((((((((...(((.(((......))).......(((..((....))..))))))..)))))))))))))..... ( -31.90) >DroYak_CAF1 5128 110 - 1 UAAUAAUGGGCA--GAA---CCGUG-AGGCCAACUUCUUU---GAGUUUGUCUU-UCCCACGCUAAACUGCGAAACCAAAUGUGAGAAAAUUUGCAUGGGUCAGGCGGAUUGGGAACGUA .....((((...--...---))))(-(((....))))(((---.(((((((((.-.(((.(((......))).......(((..((....))..))))))..))))))))).)))..... ( -28.30) >consensus CUAUUAUGGGCUAUGGG___CCAUG_AGGCCAACUUCUUU___GAGUUCGUCUUUUCCCACGCUAAACUGCGAAACCAAAUGUGAGAAAAUUUGCAUGGGUCAGGCGGAUUCGGAACGUA ...........................................((((((((((...(((.(((......))).......(((((((....))))))))))..))))))))))........ (-15.32 = -17.77 + 2.45)

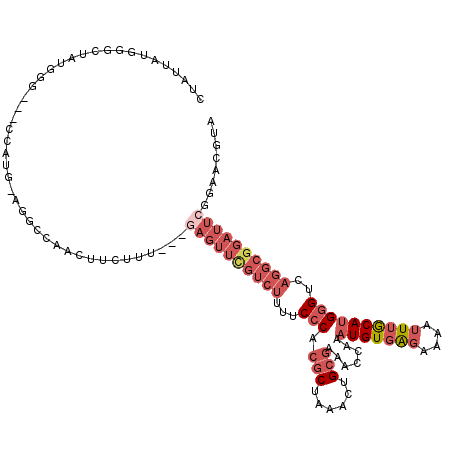
| Location | 19,470,126 – 19,470,218 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 88.16 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.34 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19470126 92 - 20766785 AGCAGUUCAAGGUUUUGGGCUAUUAUGGACUAUGGGCCAUGAGGCCAACUUCUUUGAGUUCGUCUUUUCCCACGCUAAACUGCGAAAUUGAA .((((((...(((..((((..(..(((((((...((((....))))..(......))))))))..)..)))).))).))))))......... ( -27.30) >DroSec_CAF1 4808 91 - 1 AGCAGUUCAAGGUU-UGGGCUAUUAUGGACUAUGGGCCAUGGGCCCAACUUCUUUGAGUUCGUCUUUUCGCACGCUAAACUGCGAAACCGAA .....(((((((.(-((((((..(((((.(.....)))))))))))))...)))))))((((...(((((((........))))))).)))) ( -29.60) >DroSim_CAF1 6055 91 - 1 AGCAGUUCAAGGUU-UGGGCUAUUAUGGGCUAUGGGCCAUGAGGCCAACUUCUUUGAGUUCGUCUUUUCCCACGCUAAACUGCGAAACCGAA .((((((...(((.-((((...((((((.(....).))))))(((.(((((....))))).)))....)))).))).))))))......... ( -28.90) >DroEre_CAF1 4732 89 - 1 UGCAGUUCAAGGUU-UGGGCAAUUAUUGGCA--GAACCAUGAGGCCAACUUCUUUGAGUUCGUCUUUUCCCACGCUAAACUGCGAAAUCAAA .((((((...(((.-((((.((.....(((.--((((((.((((....))))..)).)))))))..)))))).))).))))))......... ( -24.70) >DroYak_CAF1 5168 88 - 1 UGCAGUUCAAGGUU-UUGGUAAUAAUGGGCA--GAACCGUGAGGCCAACUUCUUUGAGUUUGUCUU-UCCCACGCUAAACUGCGAAACCAAA .((((((...((((-(((.(.......).))--)))))(((((((.(((((....))))).)))).-...)))....))))))......... ( -23.80) >consensus AGCAGUUCAAGGUU_UGGGCUAUUAUGGGCUAUGGGCCAUGAGGCCAACUUCUUUGAGUUCGUCUUUUCCCACGCUAAACUGCGAAACCGAA .(((((((((((.....((((.((((((........)))))))))).....))))))...(((........)))....)))))......... (-19.94 = -20.34 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:56 2006