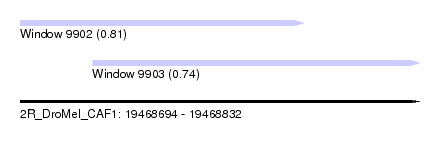
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,468,694 – 19,468,832 |
| Length | 138 |
| Max. P | 0.813086 |

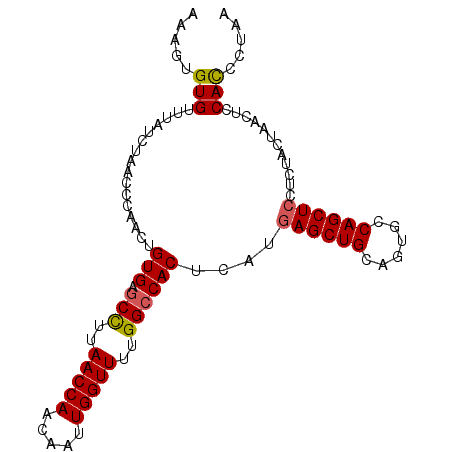
| Location | 19,468,694 – 19,468,792 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.80 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -19.17 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.813086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19468694 98 + 20766785 AUCGUGUGUUUAUCUAACCCAACUGUGACCUUUAACCAACAAUUGGUUUUGGCCACUCAUAAGCUGCAGUGCCAGCUCCUCUACGAACUCCAUCCUAA .(((((..................(((.((...(((((.....)))))..)).))).....(((((......)))))....)))))............ ( -19.10) >DroSec_CAF1 3384 98 + 1 AAAAAGUGUUUAUCUAACCCAACUGUGAGCCUUAACCAACGAUUGGUUUUGGCCACUCAUGAGCUGCAGUGCCAGCUCCUCUACUAACUCCACCCUAA .....(((................(((.(((..(((((.....)))))..))))))....((((((......))))))............)))..... ( -24.40) >DroSim_CAF1 4034 98 + 1 AAAGUGUGUUUAUCUAGCCCAACUGUGAGCCUUAACCAACAAUUGGUUUUGGCCACCUAUGAGCUGCAGAGCCAGCUCCUCUACUUACUCCACCCUAA ...(((.(..((..(((.......(((.(((..(((((.....)))))..))))))....((((((......))))))..)))..))..))))..... ( -25.60) >consensus AAAGUGUGUUUAUCUAACCCAACUGUGAGCCUUAACCAACAAUUGGUUUUGGCCACUCAUGAGCUGCAGUGCCAGCUCCUCUACUAACUCCACCCUAA .....(((................(((.(((..(((((.....)))))..))))))....((((((......))))))............)))..... (-19.17 = -19.40 + 0.23)

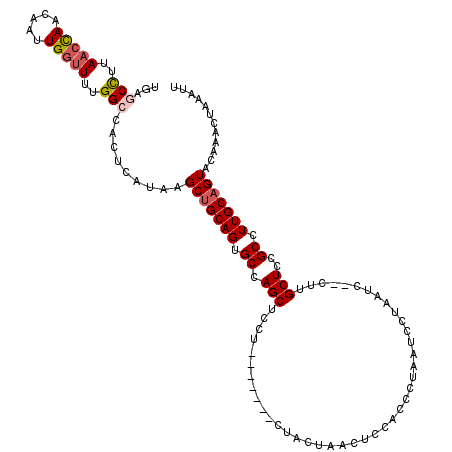
| Location | 19,468,719 – 19,468,832 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.27 |
| Mean single sequence MFE | -19.84 |
| Consensus MFE | -14.84 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19468719 113 + 20766785 UGACCUUUAACCAACAAUUGGUUUUGGCCACUCAUAAGCUGCAGUGCCAGCUCCU-------CUACGAACUCCAUCCUAAUCCUAAUCCCCUUGCUCCGCCUUGCAGUACAAACUAAAUU ((.((...(((((.....)))))..)).)).......(((((((.((.(((....-------...............................)))..)).)))))))............ ( -18.55) >DroSec_CAF1 3409 111 + 1 UGAGCCUUAACCAACGAUUGGUUUUGGCCACUCAUGAGCUGCAGUGCCAGCUCCU-------CUACUAACUCCACCCUAAUCCUAAUC--CUUGCUCCGCCUUGCAGUACAAACUAAAUU ((.(((..(((((.....)))))..)))))....((.(((((((.((.(((....-------..........................--...)))..)).))))))).))......... ( -24.31) >DroSim_CAF1 4059 111 + 1 UGAGCCUUAACCAACAAUUGGUUUUGGCCACCUAUGAGCUGCAGAGCCAGCUCCU-------CUACUUACUCCACCCUAAUCCUAAUC--CUUGCUCCGCCUUGCAGUACAAAAUAAAUU ((.(((..(((((.....)))))..)))))....((.(((((((.((.(((....-------..........................--...)))..)).))))))).))......... ( -24.31) >DroYak_CAF1 3752 111 + 1 -GAACUUUAACUACCAAUUGACUUUGGCCACUCGUAAGCCGCAGUGCCAGCCCCUACUACUCCUACUAACACCACCCUAAU------C--CUUGCUCCGCCUUGCAGUACAAACUAAAUU -................(((((.(((((.(((((.....)).))))))))...............................------.--..(((........))))).)))........ ( -12.20) >consensus UGAGCCUUAACCAACAAUUGGUUUUGGCCACUCAUAAGCUGCAGUGCCAGCUCCU_______CUACUAACUCCACCCUAAUCCUAAUC__CUUGCUCCGCCUUGCAGUACAAACUAAAUU ...(((..(((((.....)))))..))).........(((((((.((.(((..........................................)))..)).)))))))............ (-14.84 = -15.65 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:54 2006