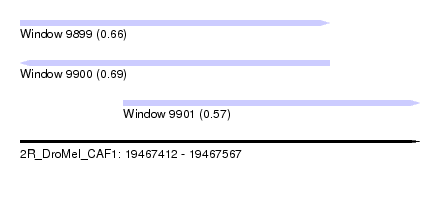
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,467,412 – 19,467,567 |
| Length | 155 |
| Max. P | 0.691114 |

| Location | 19,467,412 – 19,467,532 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -45.90 |
| Consensus MFE | -31.08 |
| Energy contribution | -30.92 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
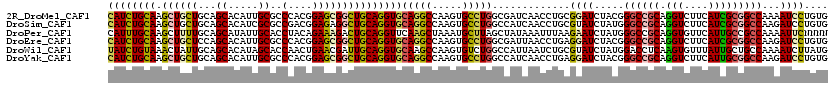
>2R_DroMel_CAF1 19467412 120 + 20766785 CAUCUGCAAGCUGCUGCAGCACAUUGCGCCCACGGAGCGGCUGCAGGUGCAGGCCAAGUGCCUGGCGAUCAACCUGCGGAUCUACGGGCCGCAGGUCUUCAUCGCGGCCAAAAUCCUGUG .(((((((.((((((.(.((.....))(....)).)))))))))))))(((((......(((..(((((..((((((((.((....))))))))))....))))))))......))))). ( -55.40) >DroSim_CAF1 2754 120 + 1 CAUCUGCAAGCUGCUGCAGCACAUCGCGCCGACGGAGAGGCUGCAGGUGCAGGCCAAGUGCCUGGCCAUCAACCUGCGUAUCUAUGGGCCGCAGGUCUUCAUCGCGGCCAAGAUCCUGUG ...(((((......)))))((((..(((((........))).((((((...(((((......)))))....))))))))((((...((((((.(((....))))))))).))))..)))) ( -47.30) >DroPer_CAF1 3800 120 + 1 CAUUUGCAAGCUUUUGCAGCAUAUUGCACCUACAGAAAGACUGCAGGUUCAAGCUAAAUGCUUAGCUAUAAAUUUAAGAAUCUAUGGGCCGCAGGUGUUCAUUGCCGCCAAAAUUCNNNN ...((((((....))))))((((.((.((((.(((.....))).)))).))((((((....))))))...............))))(((.((((.......)))).)))........... ( -27.10) >DroEre_CAF1 2074 120 + 1 CAUCUGCAAGCUGCUCCAGCACAUUGCGCCCACGGAGCGGCUGCAGGUGCAGGCCAAGUGCCUGGCGAUUAACCUGAGGAUCUACGGGCCGCAGGUCUUCAUCGCGGCCAAGAUCCUGUG ((((((((.((((((((.((.....))(....)))))))))))))))))(((((.....)))))............(((((((...((((((.(((....))))))))).)))))))... ( -59.00) >DroWil_CAF1 1480 120 + 1 UAUCUGUAAACUAUUGCAGCACAUAGCACCAACUGAACGAUUGCAGGUGCAAGCCAAGUGUCUGGCCAUUAAUCUGCGUAUCUAUGGACCUCAAGUGUUUAUUGCUGCCAAAAUCUUAUG ...............((((((...(((((.........((((((((((....((((......)))).....))))))).)))..((.....)).)))))...))))))............ ( -29.10) >DroYak_CAF1 2402 120 + 1 CAUCUGCAAGCUGCUGCAGCACAUUGCGCCCACGGAGCGGCUGCAGGUGCAGGCCAAGUGCCUGGCCAUCAACCUGAGGAUCUACGGGCCGCAGGUCUUCAUUGCGGCCAAGAUCCUGUG ((((((((.((((((.(.((.....))(....)).))))))))))))))..(((((......))))).........(((((((...((((((((.......)))))))).)))))))... ( -57.50) >consensus CAUCUGCAAGCUGCUGCAGCACAUUGCGCCCACGGAGCGGCUGCAGGUGCAGGCCAAGUGCCUGGCCAUCAACCUGAGGAUCUACGGGCCGCAGGUCUUCAUCGCGGCCAAAAUCCUGUG ((((((((.((((((...((.....))..(....)))))))))))))))(((((.....))))).............((((.....((((((.(((....)))))))))...)))).... (-31.08 = -30.92 + -0.16)
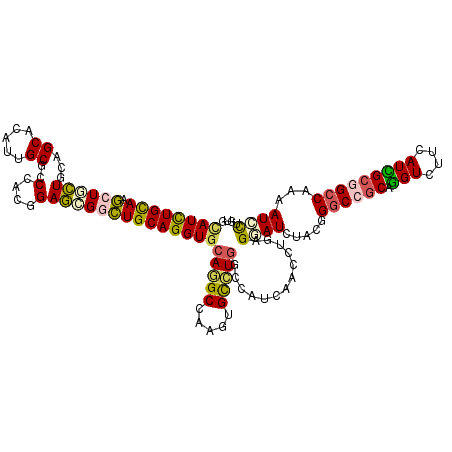
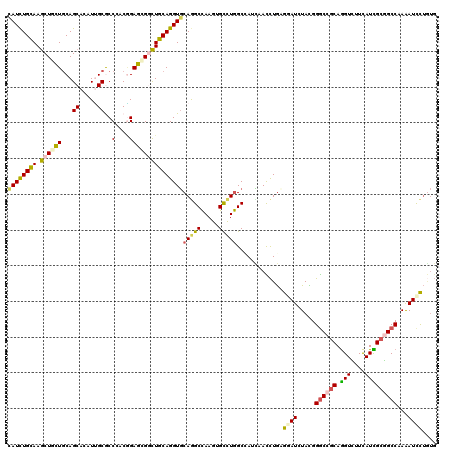
| Location | 19,467,412 – 19,467,532 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -46.53 |
| Consensus MFE | -34.11 |
| Energy contribution | -33.48 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19467412 120 - 20766785 CACAGGAUUUUGGCCGCGAUGAAGACCUGCGGCCCGUAGAUCCGCAGGUUGAUCGCCAGGCACUUGGCCUGCACCUGCAGCCGCUCCGUGGGCGCAAUGUGCUGCAGCAGCUUGCAGAUG ....((((((.(((((((.........)))))))...))))))((((((((...((.((((.....))))))..(((((((((((.....))))......)))))))))))))))..... ( -55.10) >DroSim_CAF1 2754 120 - 1 CACAGGAUCUUGGCCGCGAUGAAGACCUGCGGCCCAUAGAUACGCAGGUUGAUGGCCAGGCACUUGGCCUGCACCUGCAGCCUCUCCGUCGGCGCGAUGUGCUGCAGCAGCUUGCAGAUG ....(.((((.(((((((.........)))))))...)))).)((((((((..((((((....)))))).....(((((((...((((....)).))...)))))))))))))))..... ( -50.60) >DroPer_CAF1 3800 120 - 1 NNNNGAAUUUUGGCGGCAAUGAACACCUGCGGCCCAUAGAUUCUUAAAUUUAUAGCUAAGCAUUUAGCUUGAACCUGCAGUCUUUCUGUAGGUGCAAUAUGCUGCAAAAGCUUGCAAAUG ....((((((.(((.(((.........))).)))...))))))...........((.((((.(((((((((.((((((((.....)))))))).)))...))))..)).))))))..... ( -34.40) >DroEre_CAF1 2074 120 - 1 CACAGGAUCUUGGCCGCGAUGAAGACCUGCGGCCCGUAGAUCCUCAGGUUAAUCGCCAGGCACUUGGCCUGCACCUGCAGCCGCUCCGUGGGCGCAAUGUGCUGGAGCAGCUUGCAGAUG ...(((((((.(((((((.........)))))))...)))))))((((((((..((...))..))))))))...(((((((.(((((...(((((...)))))))))).)).)))))... ( -54.00) >DroWil_CAF1 1480 120 - 1 CAUAAGAUUUUGGCAGCAAUAAACACUUGAGGUCCAUAGAUACGCAGAUUAAUGGCCAGACACUUGGCUUGCACCUGCAAUCGUUCAGUUGGUGCUAUGUGCUGCAAUAGUUUACAGAUA ....(((((...((((((.....(((((((((((....)))..((((......((((((....)))))).....)))).....))))...)))).....))))))...)))))....... ( -30.40) >DroYak_CAF1 2402 120 - 1 CACAGGAUCUUGGCCGCAAUGAAGACCUGCGGCCCGUAGAUCCUCAGGUUGAUGGCCAGGCACUUGGCCUGCACCUGCAGCCGCUCCGUGGGCGCAAUGUGCUGCAGCAGCUUGCAGAUG ...(((((((.(((((((.........)))))))...)))))))(((((((..((((((....)))))).....(((((((((((.....))))......))))))))))))))...... ( -54.70) >consensus CACAGGAUCUUGGCCGCAAUGAAGACCUGCGGCCCAUAGAUCCGCAGGUUGAUGGCCAGGCACUUGGCCUGCACCUGCAGCCGCUCCGUGGGCGCAAUGUGCUGCAGCAGCUUGCAGAUG ....((((((.(((((((.........)))))))...)))))).(((((((..((((((....)))))).((((.(((.(((........))))))..)))).....)))))))...... (-34.11 = -33.48 + -0.63)

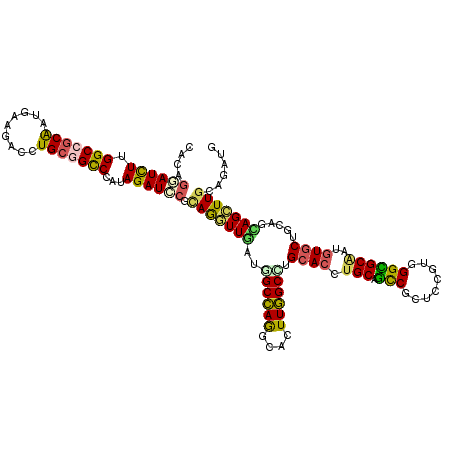
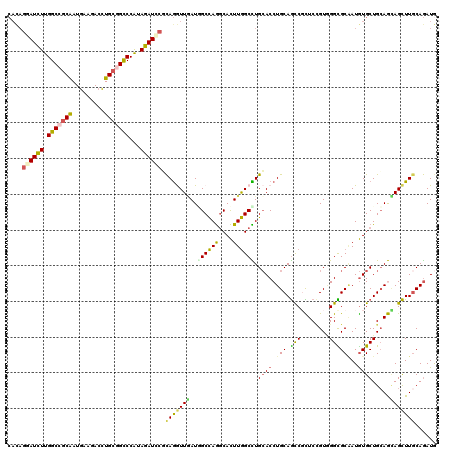
| Location | 19,467,452 – 19,467,567 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -44.93 |
| Consensus MFE | -35.71 |
| Energy contribution | -36.88 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19467452 115 + 20766785 CUGCAGGUGCAGGCCAAGUGCCUGGCGAUCAACCUGCGGAUCUACGGGCCGCAGGUCUUCAUCGCGGCCAAAAUCCUGUGGAAGUACUCCAACAGCAGUGCGGCCAACGAGAACU ...((((..(.......)..))))(((((..((((((((.((....))))))))))....)))))((((......(((((((.....))).))))......)))).......... ( -48.40) >DroSec_CAF1 2100 115 + 1 CUGCAGGUGCAGGCCAAGUGCCUGGCCAUCAACCUGCGGAUCUACGGGCCGCAGGUCUUCAUCGCGGCCAAGAUCCUGUGGAAGUACUCCAACAGCAGUGCGGCCAACGAGAACU ((((((((...(((((......)))))....)))))))).(((...(((((((((((........))))......(((((((.....))).))))...)))))))....)))... ( -51.30) >DroSim_CAF1 2794 115 + 1 CUGCAGGUGCAGGCCAAGUGCCUGGCCAUCAACCUGCGUAUCUAUGGGCCGCAGGUCUUCAUCGCGGCCAAGAUCCUGUGGAAGUACUCCAACAGCAGUGCGGCCAACGAGAACU ..((((((...(((((......)))))....))))))((.(((...(((((((((((........))))......(((((((.....))).))))...)))))))....))))). ( -48.60) >DroEre_CAF1 2114 115 + 1 CUGCAGGUGCAGGCCAAGUGCCUGGCGAUUAACCUGAGGAUCUACGGGCCGCAGGUCUUCAUCGCGGCCAAGAUCCUGUGGAAGUACUCCAACAGCAGUGCGGCCAACGAGAACU ...(((((....((((......)))).....)))))(((((((...((((((.(((....))))))))).))))))).(((..(((((.(....).)))))..)))......... ( -44.90) >DroWil_CAF1 1520 115 + 1 UUGCAGGUGCAAGCCAAGUGUCUGGCCAUUAAUCUGCGUAUCUAUGGACCUCAAGUGUUUAUUGCUGCCAAAAUCUUAUGGAAAUAUUCAAAUAGCAGUGCAGCAAAUGAAAAUU ..((((((....((((......)))).....)))))).............(((..(((((((((((((((........)))...........)))))))).))))..)))..... ( -25.90) >DroYak_CAF1 2442 115 + 1 CUGCAGGUGCAGGCCAAGUGCCUGGCCAUCAACCUGAGGAUCUACGGGCCGCAGGUCUUCAUUGCGGCCAAGAUCCUGUGGAAGUACUCGAACAGCAGUGCGGCCAACGAGAACU ...(((((...(((((......)))))....)))))(((((((...((((((((.......)))))))).))))))).(((..((((((.....).)))))..)))......... ( -50.50) >consensus CUGCAGGUGCAGGCCAAGUGCCUGGCCAUCAACCUGCGGAUCUACGGGCCGCAGGUCUUCAUCGCGGCCAAGAUCCUGUGGAAGUACUCCAACAGCAGUGCGGCCAACGAGAACU ((((((((....((((......)))).....)))))))).(((...(((((((((((........))))......(((((((.....))).))))...)))))))....)))... (-35.71 = -36.88 + 1.17)

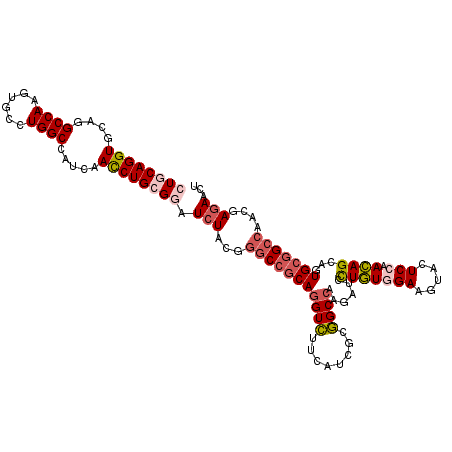
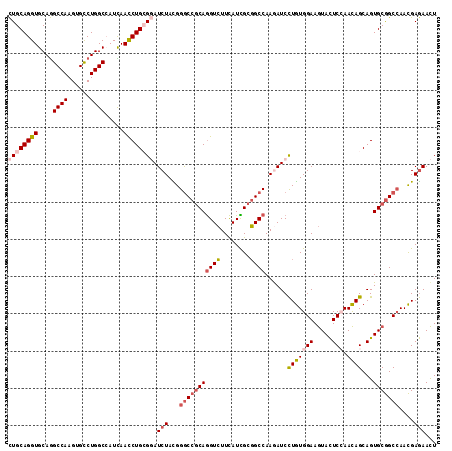
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:53 2006