
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
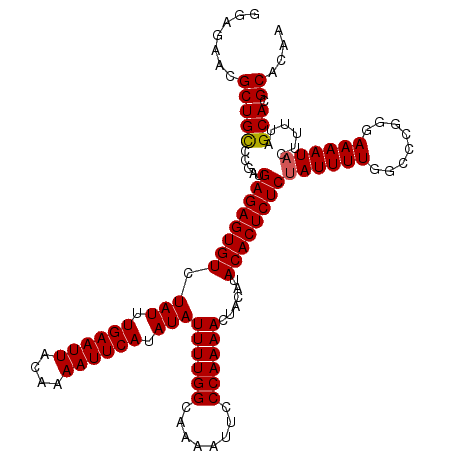
| Location | 19,443,288 – 19,443,408 |
| Length | 120 |
| Max. P | 0.980078 |

| Location | 19,443,288 – 19,443,408 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -24.39 |
| Energy contribution | -24.42 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
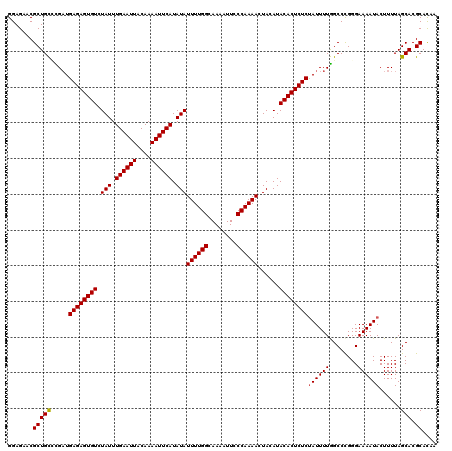
>2R_DroMel_CAF1 19443288 120 + 20766785 GGAGAACGCUGCGCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUAUUUUGGCCCGGGAAAAUACUUUUAGCACGCACAA .......(((((.....((((((((.(((.((((((....)))))).)))((((((........))))))......))))))))((((((........))))))......))).)).... ( -25.60) >DroGri_CAF1 14434 120 + 1 GGAGAACGCUGCUUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAAUUAGAUACACUCUCUAUUUUGGCCUGGGAAAAUACUUUUAGCACGCACAA .......((((((.((.(((((((((((..((((((....))))))...(((((((........)))))))))))..)))))))((((((........))))))..)).)))).)).... ( -26.30) >DroSec_CAF1 7794 120 + 1 GGAGAACGCUGCGCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCCAUUUUGGCCCGGGAAAAUACUUUUAGCACGCACAA .........((.(((..((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).......((..(((......)))...)).))).)). ( -24.50) >DroWil_CAF1 6956 120 + 1 GGAGAACGCUGUCUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUAUUUUAGCCCGGGAAAAUACUUUUAGCACGCACAA .......((((.((((.((((((((.(((.((((((....)))))).)))((((((........))))))......))))))))((((((........))))))...)))))).)).... ( -24.50) >DroMoj_CAF1 12802 120 + 1 GGAGAACGCUGCCUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUAGAUACACUCUCUAUUUUGGCCUGGGAAAAUACUUUUAGCACGCAUCA .......((.(((.(((((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).)))..)))((((((......))))))...)).... ( -27.30) >DroAna_CAF1 7005 120 + 1 GGAGAACGCUGCUCGUUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUAUUUUGGCCCGGGAAAAUACUUUUAGCACGCACAA .......((((((....((((((((.(((.((((((....)))))).)))((((((........))))))......))))))))((((((........)))))).....)))).)).... ( -25.60) >consensus GGAGAACGCUGCCCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUAUUUUGGCCCGGGAAAAUACUUUUAGCACGCACAA .......(((((.....((((((((.(((.((((((....)))))).)))((((((........))))))......))))))))((((((........))))))......))).)).... (-24.39 = -24.42 + 0.03)

| Location | 19,443,288 – 19,443,408 |
|---|---|
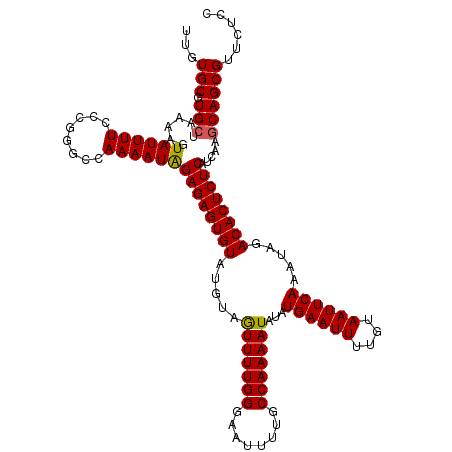
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -27.29 |
| Energy contribution | -27.18 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19443288 120 - 20766785 UUGUGCGUGCUAAAAGUAUUUUCCCGGGCCAAAAUAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCGCGCAGCGUUCUCC (((((((.(((....(......)...))).......((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))..)))))))........ ( -31.60) >DroGri_CAF1 14434 120 - 1 UUGUGCGUGCUAAAAGUAUUUUCCCAGGCCAAAAUAGAGAGUGUAUCUAAUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAAGCAGCGUUCUCC ....((.((((.....((((((........))))))(((((((..(((((((((((........)))))))...((((((....))))))..)))))))))))....))))))....... ( -29.40) >DroSec_CAF1 7794 120 - 1 UUGUGCGUGCUAAAAGUAUUUUCCCGGGCCAAAAUGGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCGCGCAGCGUUCUCC (((((((.(((....(......)...))).......((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))..)))))))........ ( -31.40) >DroWil_CAF1 6956 120 - 1 UUGUGCGUGCUAAAAGUAUUUUCCCGGGCUAAAAUAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAGACAGCGUUCUCC ..(.((((.((.....((((((........))))))((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))...))...)))).)... ( -27.30) >DroMoj_CAF1 12802 120 - 1 UGAUGCGUGCUAAAAGUAUUUUCCCAGGCCAAAAUAGAGAGUGUAUCUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAGGCAGCGUUCUCC .(((((.((((.....((((((........))))))(((((((..(((((((((((........)))))))...((((((....))))))..)))))))))))....))))))))).... ( -31.90) >DroAna_CAF1 7005 120 - 1 UUGUGCGUGCUAAAAGUAUUUUCCCGGGCCAAAAUAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAACGAGCAGCGUUCUCC ....((.((((.....((((((........))))))((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))....))))))....... ( -30.10) >consensus UUGUGCGUGCUAAAAGUAUUUUCCCGGGCCAAAAUAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAAGCAGCGUUCUCC ...(((.(((......((((((........))))))((((((((.....(((((((........)))))))...((((((....)))))).....)))))))).....))))))...... (-27.29 = -27.18 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:45 2006