| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,420,179 – 19,420,269 |
| Length | 90 |
| Max. P | 0.655448 |

| Location | 19,420,179 – 19,420,269 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.27 |
| Mean single sequence MFE | -30.98 |
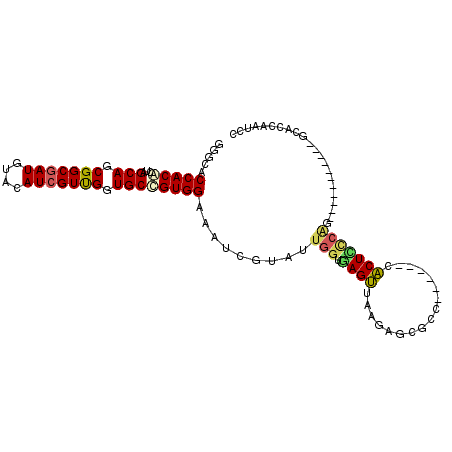
| Consensus MFE | -20.50 |
| Energy contribution | -20.53 |
| Covariance contribution | 0.03 |
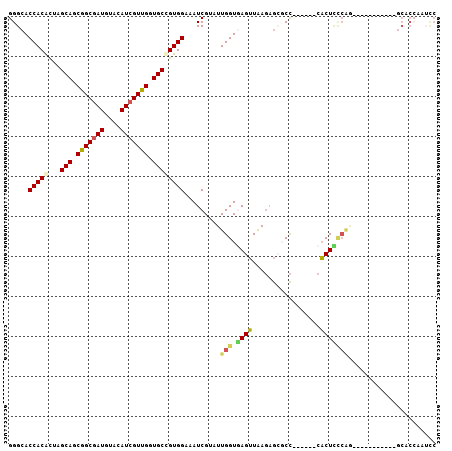
| Combinations/Pair | 1.32 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
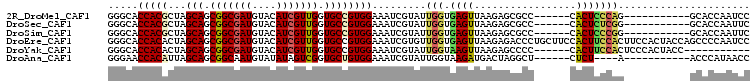
>2R_DroMel_CAF1 19420179 90 + 20766785 GGGCACCACGCUAGCAGCGGCGAUGUACAUCGUUGGUGCCGUGGAAAUCGUAUUGGUGAGUUAAGAGCGCC------CACUCCCAG-----------GCACCAAUCC ((((.(((((...(((.(((((((....))))))).))))))))........((((.((((..........------.))))))))-----------)).))..... ( -32.10) >DroSec_CAF1 2850 90 + 1 GGGCACCACGCUAGCAGCGGCGAUGUACAUCGUUGGUGCCGUGGAAAUCGUAUUGGUGAGUUAAGAGCGCC------CACUCUCGG-----------GCACCAAUUC .((((((((((.....)))(((((....))))))))))))...........((((((..((.....))(((------(......))-----------)))))))).. ( -34.20) >DroSim_CAF1 2872 90 + 1 GGGCACCACGCUAGCAGCGGCGAUGUACAUCGUUGGUGCCGUGGAAAUCGUAUUGGUGAGUUAAGAGCGCC------CACUCCCGG-----------GCACCAAUUC .((((((((((.....)))(((((....))))))))))))...........((((((..((.....))(((------(......))-----------)))))))).. ( -34.20) >DroEre_CAF1 2976 107 + 1 GGGCACCACACUAGCAGCGGCGAUGUACAUCGUUGGUGCCGUGGAAAUCGUGUUGGUGAGUUAAGAGACCCUGCUUCCACUUCCACUUCCACUACCAGCCCCAAUCC ((((.........(((.(((((((....))))))).))).((((((...(((..((((.(...((.....))....)))))..))))))))).....))))...... ( -36.80) >DroYak_CAF1 3016 90 + 1 GGGCACCACACUAGCAGCGGCGAUGUACAUCGUUGGUGCCGUGGAAAUCGUAUUGGUAAGUUAAGAGCCCC------CACUUCCACUCCCACUACC----------- ((((.((((....(((.(((((((....))))))).))).))))...((..(((....)))...)))))).------...................----------- ( -26.00) >DroAna_CAF1 2844 86 + 1 GGGAACCACAUUAGCAGCGGCAAUGUAUAUAGUCGGUGCUGUGGAAAUCGUAUUGGUAAGAUGACUAGGCU------CUCU----A-----------ACCCAUAACC (((..(((((...(((.((((..........)))).)))))))).....((.(((((......))))))).------....----.-----------.)))...... ( -22.60) >consensus GGGCACCACACUAGCAGCGGCGAUGUACAUCGUUGGUGCCGUGGAAAUCGUAUUGGUGAGUUAAGAGCGCC______CACUCCCAG___________GCACCAAUCC .....(((((...(((.(((((((....))))))).)))))))).........(((.((((.................)))))))...................... (-20.50 = -20.53 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:39 2006